Probe CUST_41424_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41424_PI426222305 | JHI_St_60k_v1 | DMT400079508 | GTCAAATCACTTGGACTTGTTGCAATATGTTAATCTGAATGGGAATGGGTTTTAACAGAT |
All Microarray Probes Designed to Gene DMG400030958
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41381_PI426222305 | JHI_St_60k_v1 | DMT400079507 | CTCTTCTTCTTCATATGATGAAGATTCAATGTGATGTTTGTGATAAAGAAGAGGCATCAG |
| CUST_41424_PI426222305 | JHI_St_60k_v1 | DMT400079508 | GTCAAATCACTTGGACTTGTTGCAATATGTTAATCTGAATGGGAATGGGTTTTAACAGAT |
Microarray Signals from CUST_41424_PI426222305
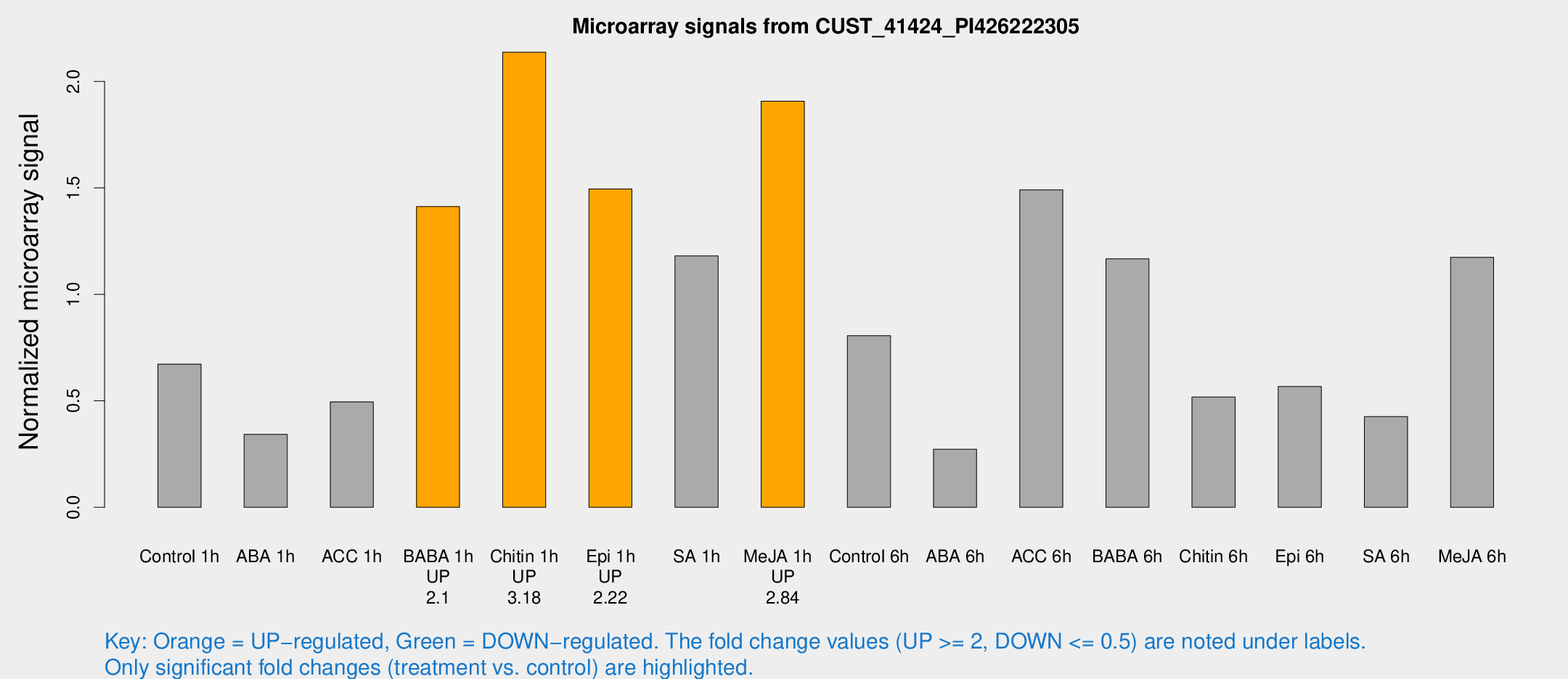
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.4739 | 3.5661 | 0.672717 | 0.113022 |
| ABA 1h | 10.0697 | 3.14626 | 0.342523 | 0.128091 |
| ACC 1h | 19.4573 | 7.45927 | 0.495369 | 0.314869 |
| BABA 1h | 44.3489 | 9.40372 | 1.41247 | 0.188957 |
| Chitin 1h | 61.0625 | 8.7361 | 2.13734 | 0.514202 |
| Epi 1h | 40.3841 | 3.96989 | 1.495 | 0.146862 |
| SA 1h | 38.1042 | 3.90073 | 1.18071 | 0.14945 |
| Me-JA 1h | 48.8921 | 4.87952 | 1.90725 | 0.17272 |
| Control 6h | 35.1872 | 14.3766 | 0.806119 | 0.623059 |
| ABA 6h | 9.27705 | 3.68705 | 0.272994 | 0.114158 |
| ACC 6h | 53.6444 | 5.2536 | 1.49126 | 0.184733 |
| BABA 6h | 45.5821 | 15.6343 | 1.16713 | 0.409095 |
| Chitin 6h | 19.756 | 7.60988 | 0.518262 | 0.208809 |
| Epi 6h | 20.1853 | 4.13069 | 0.56723 | 0.125086 |
| SA 6h | 13.9155 | 3.76474 | 0.425811 | 0.13217 |
| Me-JA 6h | 42.0551 | 15.4867 | 1.17435 | 0.384898 |
Source Transcript PGSC0003DMT400079508 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G39070.1 | +2 | 5e-71 | 229 | 120/203 (59%) | B-box zinc finger family protein | chr4:18205061-18206421 REVERSE LENGTH=242 |
