Probe CUST_39140_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39140_PI426222305 | JHI_St_60k_v1 | DMT400036326 | GCTTTCTCAGGAATGCTTTTTTAGTGTTGAGCTGTGTTTGTCTTGTCCTTGTAAATAATT |
All Microarray Probes Designed to Gene DMG400013973
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39155_PI426222305 | JHI_St_60k_v1 | DMT400036325 | GTAGAGCACGGAAGAGGTTCGGCGATGGTTACTGGTAACACTGCTTCACTTTTCTTTTTT |
| CUST_39140_PI426222305 | JHI_St_60k_v1 | DMT400036326 | GCTTTCTCAGGAATGCTTTTTTAGTGTTGAGCTGTGTTTGTCTTGTCCTTGTAAATAATT |
Microarray Signals from CUST_39140_PI426222305
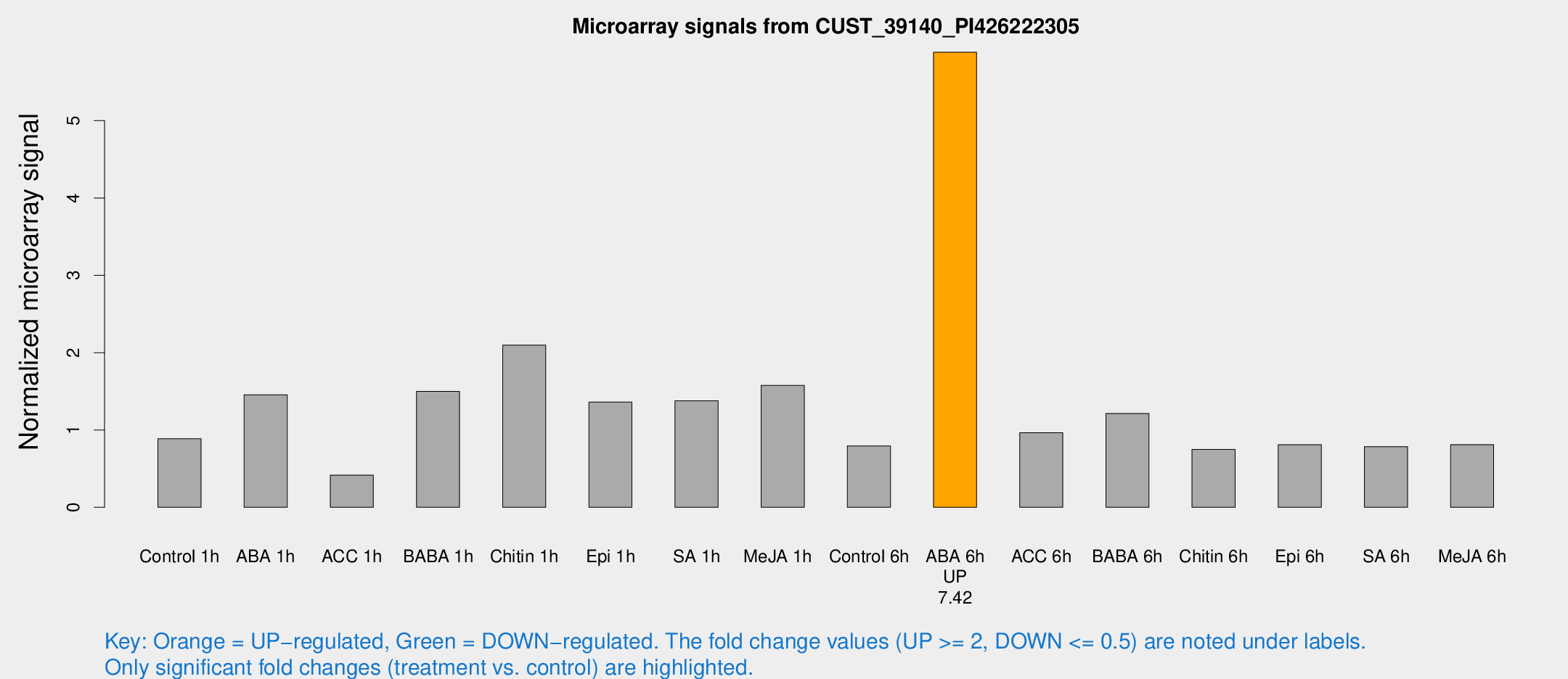
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 139.618 | 24.3064 | 0.886577 | 0.214665 |
| ABA 1h | 220.672 | 73.009 | 1.45528 | 0.415441 |
| ACC 1h | 69.4152 | 15.3925 | 0.415896 | 0.0772807 |
| BABA 1h | 247.501 | 77.0435 | 1.49851 | 0.39745 |
| Chitin 1h | 290.555 | 24.0932 | 2.09694 | 0.291491 |
| Epi 1h | 190.803 | 40.96 | 1.36168 | 0.327222 |
| SA 1h | 233.484 | 62.5153 | 1.37782 | 0.369472 |
| Me-JA 1h | 200.793 | 28.8373 | 1.57675 | 0.127901 |
| Control 6h | 131.222 | 36.592 | 0.792713 | 0.179127 |
| ABA 6h | 990.396 | 178.857 | 5.885 | 0.779828 |
| ACC 6h | 170.47 | 15.1557 | 0.96356 | 0.0792016 |
| BABA 6h | 216.142 | 41.3035 | 1.21227 | 0.203323 |
| Chitin 6h | 123.09 | 11.0765 | 0.749396 | 0.0583192 |
| Epi 6h | 143.574 | 22.441 | 0.809851 | 0.0769931 |
| SA 6h | 125.794 | 26.9849 | 0.784999 | 0.103884 |
| Me-JA 6h | 139.37 | 40.6454 | 0.81028 | 0.260553 |
Source Transcript PGSC0003DMT400036326 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
