Probe CUST_30001_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30001_PI426222305 | JHI_St_60k_v1 | DMT400005621 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
All Microarray Probes Designed to Gene DMG400002204
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29983_PI426222305 | JHI_St_60k_v1 | DMT400005620 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30027_PI426222305 | JHI_St_60k_v1 | DMT400005624 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30029_PI426222305 | JHI_St_60k_v1 | DMT400005622 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30016_PI426222305 | JHI_St_60k_v1 | DMT400005623 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
| CUST_30001_PI426222305 | JHI_St_60k_v1 | DMT400005621 | AAATATCTTCTTTGTAATGGTTACTGTGAGAAGTTTTGAATCCATCTGTACATGCTGGAC |
Microarray Signals from CUST_30001_PI426222305
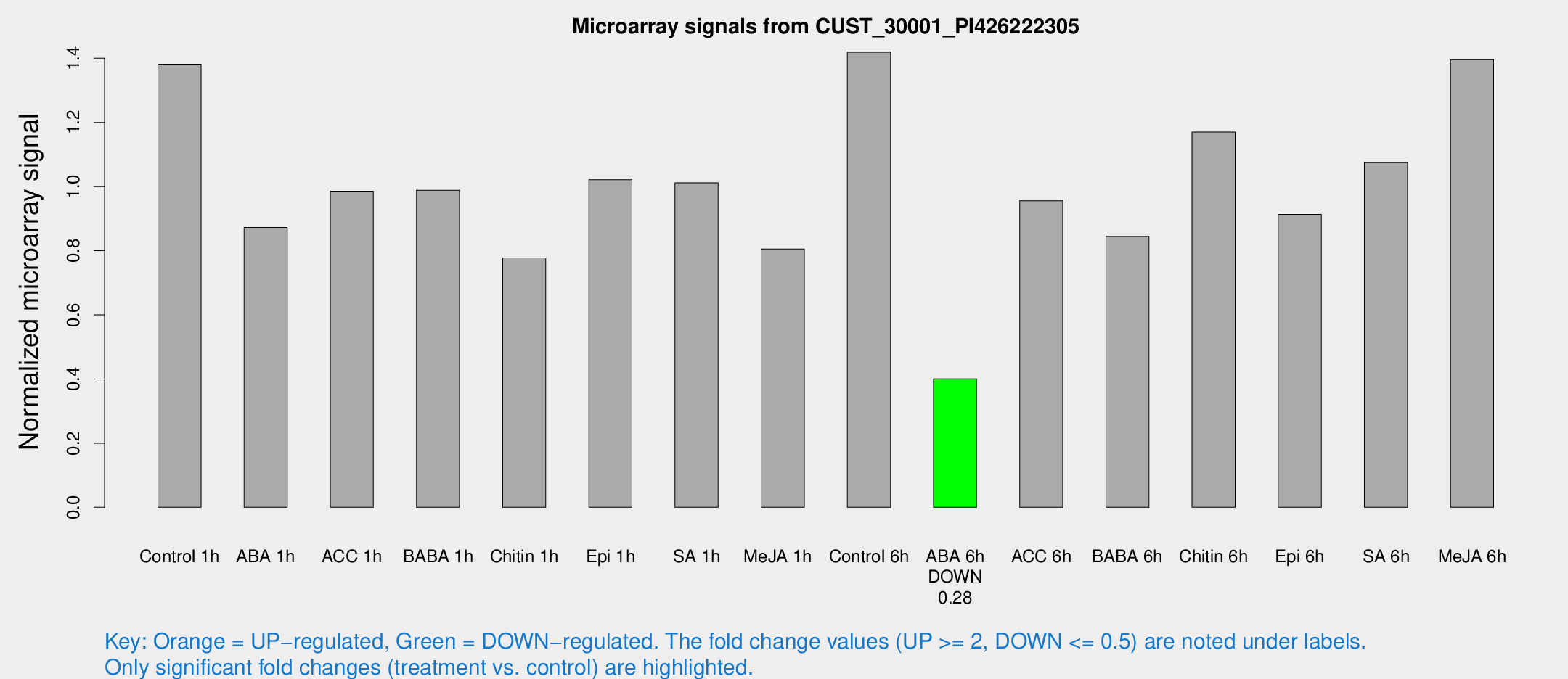
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1150.11 | 256.455 | 1.38117 | 0.217426 |
| ABA 1h | 611.978 | 35.4804 | 0.872609 | 0.0606995 |
| ACC 1h | 891.895 | 251.244 | 0.985693 | 0.278263 |
| BABA 1h | 760.256 | 58.769 | 0.988676 | 0.0572747 |
| Chitin 1h | 566.079 | 80.5729 | 0.777712 | 0.0951989 |
| Epi 1h | 701.448 | 40.6682 | 1.02125 | 0.0591503 |
| SA 1h | 850.134 | 153.542 | 1.01178 | 0.211421 |
| Me-JA 1h | 522.278 | 30.4031 | 0.805449 | 0.0697006 |
| Control 6h | 1217.07 | 295.792 | 1.41873 | 0.291975 |
| ABA 6h | 340.003 | 30.6631 | 0.400197 | 0.0277896 |
| ACC 6h | 880.526 | 99.9877 | 0.955781 | 0.0553466 |
| BABA 6h | 766.708 | 117.019 | 0.844397 | 0.111926 |
| Chitin 6h | 987.532 | 57.1593 | 1.16987 | 0.067692 |
| Epi 6h | 839.33 | 131.795 | 0.913245 | 0.232031 |
| SA 6h | 865.188 | 139.608 | 1.07437 | 0.0846746 |
| Me-JA 6h | 1111.53 | 100.49 | 1.39576 | 0.0807041 |
Source Transcript PGSC0003DMT400005621 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G56750.1 | +2 | 1e-100 | 316 | 160/215 (74%) | N-MYC downregulated-like 1 | chr5:22957986-22960606 FORWARD LENGTH=346 |
