Probe CUST_28178_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28178_PI426222305 | JHI_St_60k_v1 | DMT400004542 | AAGTCTCGTTTACCCCTTCAAATGCTCTCCTATTCCGCTTTTTTCAAATGATTGCTAAAG |
All Microarray Probes Designed to Gene DMG400001804
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28100_PI426222305 | JHI_St_60k_v1 | DMT400004541 | CAAAATTTGTGGACGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28192_PI426222305 | JHI_St_60k_v1 | DMT400004540 | CAAAATTTGTGGATGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28090_PI426222305 | JHI_St_60k_v1 | DMT400004537 | CTGCTTCATGAAAAATTTCAGACAGCAAATTAAATGGTGAAATTGCTGCAGTATGTGTCT |
| CUST_28178_PI426222305 | JHI_St_60k_v1 | DMT400004542 | AAGTCTCGTTTACCCCTTCAAATGCTCTCCTATTCCGCTTTTTTCAAATGATTGCTAAAG |
| CUST_28106_PI426222305 | JHI_St_60k_v1 | DMT400004539 | ACTCGTGATTCTCGAGTATATCGATGAGACATGGAACGAAACAGCTCCTTTGCTACCTCA |
Microarray Signals from CUST_28178_PI426222305
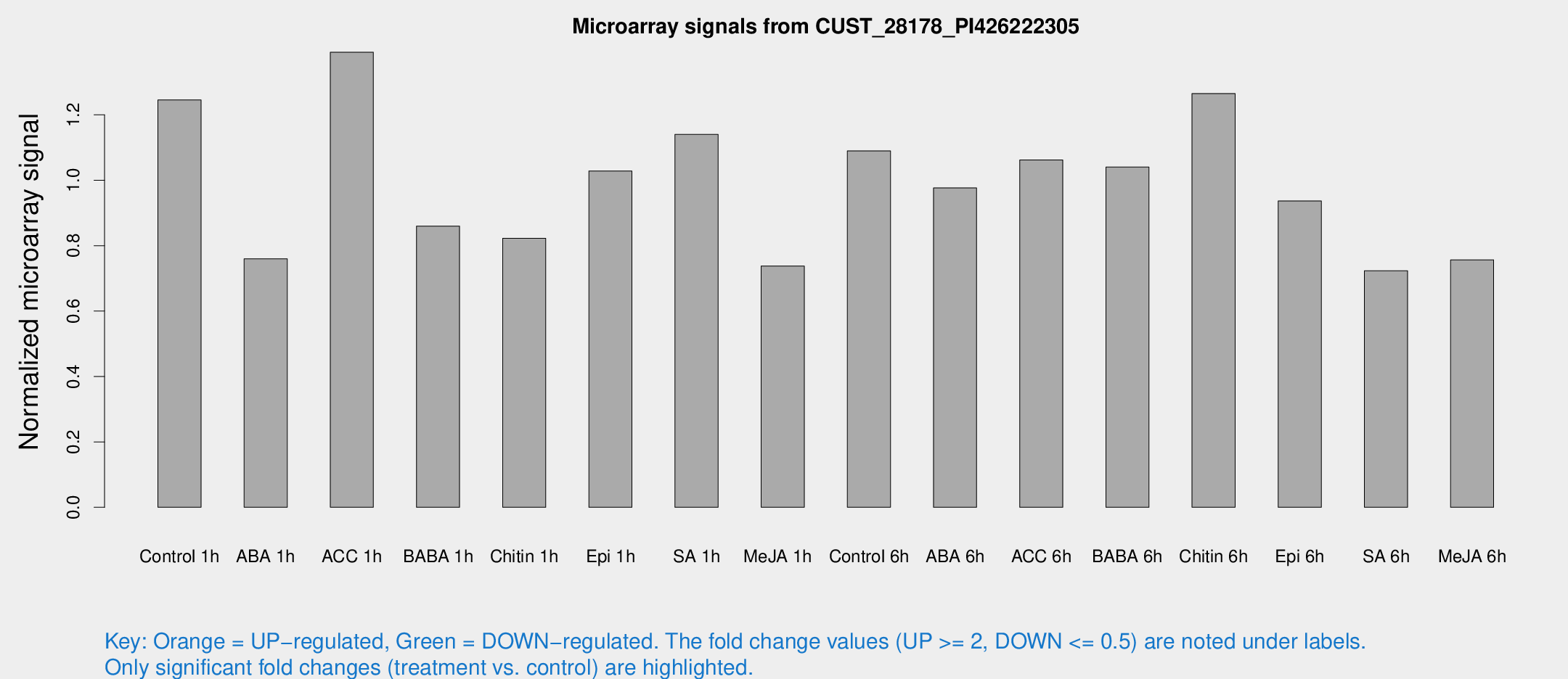
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 77.7253 | 12.4592 | 1.24576 | 0.161078 |
| ABA 1h | 46.6253 | 14.2621 | 0.759794 | 0.283972 |
| ACC 1h | 90.0763 | 16.3785 | 1.39145 | 0.179562 |
| BABA 1h | 51.9947 | 9.47966 | 0.859719 | 0.144724 |
| Chitin 1h | 47.4959 | 9.94387 | 0.822601 | 0.15421 |
| Epi 1h | 55.9921 | 10.5092 | 1.02835 | 0.164383 |
| SA 1h | 72.5344 | 10.0035 | 1.14007 | 0.100216 |
| Me-JA 1h | 39.7326 | 11.1961 | 0.737922 | 0.150386 |
| Control 6h | 76.8819 | 30.6343 | 1.08963 | 0.395832 |
| ABA 6h | 63.4772 | 4.97621 | 0.976534 | 0.117476 |
| ACC 6h | 78.1902 | 16.7463 | 1.06195 | 0.166716 |
| BABA 6h | 71.3247 | 5.94495 | 1.04018 | 0.128348 |
| Chitin 6h | 87.4528 | 21.6072 | 1.26506 | 0.299823 |
| Epi 6h | 65.8148 | 10.4301 | 0.936561 | 0.133209 |
| SA 6h | 64.1674 | 34.5204 | 0.723152 | 0.765479 |
| Me-JA 6h | 46.6698 | 6.43602 | 0.756582 | 0.0734374 |
Source Transcript PGSC0003DMT400004542 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G09270.1 | +2 | 4e-58 | 192 | 99/215 (46%) | glutathione S-transferase TAU 8 | chr3:2848407-2849226 REVERSE LENGTH=224 |
