Probe CUST_28100_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28100_PI426222305 | JHI_St_60k_v1 | DMT400004541 | CAAAATTTGTGGACGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
All Microarray Probes Designed to Gene DMG400001804
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28106_PI426222305 | JHI_St_60k_v1 | DMT400004539 | ACTCGTGATTCTCGAGTATATCGATGAGACATGGAACGAAACAGCTCCTTTGCTACCTCA |
| CUST_28192_PI426222305 | JHI_St_60k_v1 | DMT400004540 | CAAAATTTGTGGATGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28178_PI426222305 | JHI_St_60k_v1 | DMT400004542 | AAGTCTCGTTTACCCCTTCAAATGCTCTCCTATTCCGCTTTTTTCAAATGATTGCTAAAG |
| CUST_28100_PI426222305 | JHI_St_60k_v1 | DMT400004541 | CAAAATTTGTGGACGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28090_PI426222305 | JHI_St_60k_v1 | DMT400004537 | CTGCTTCATGAAAAATTTCAGACAGCAAATTAAATGGTGAAATTGCTGCAGTATGTGTCT |
Microarray Signals from CUST_28100_PI426222305
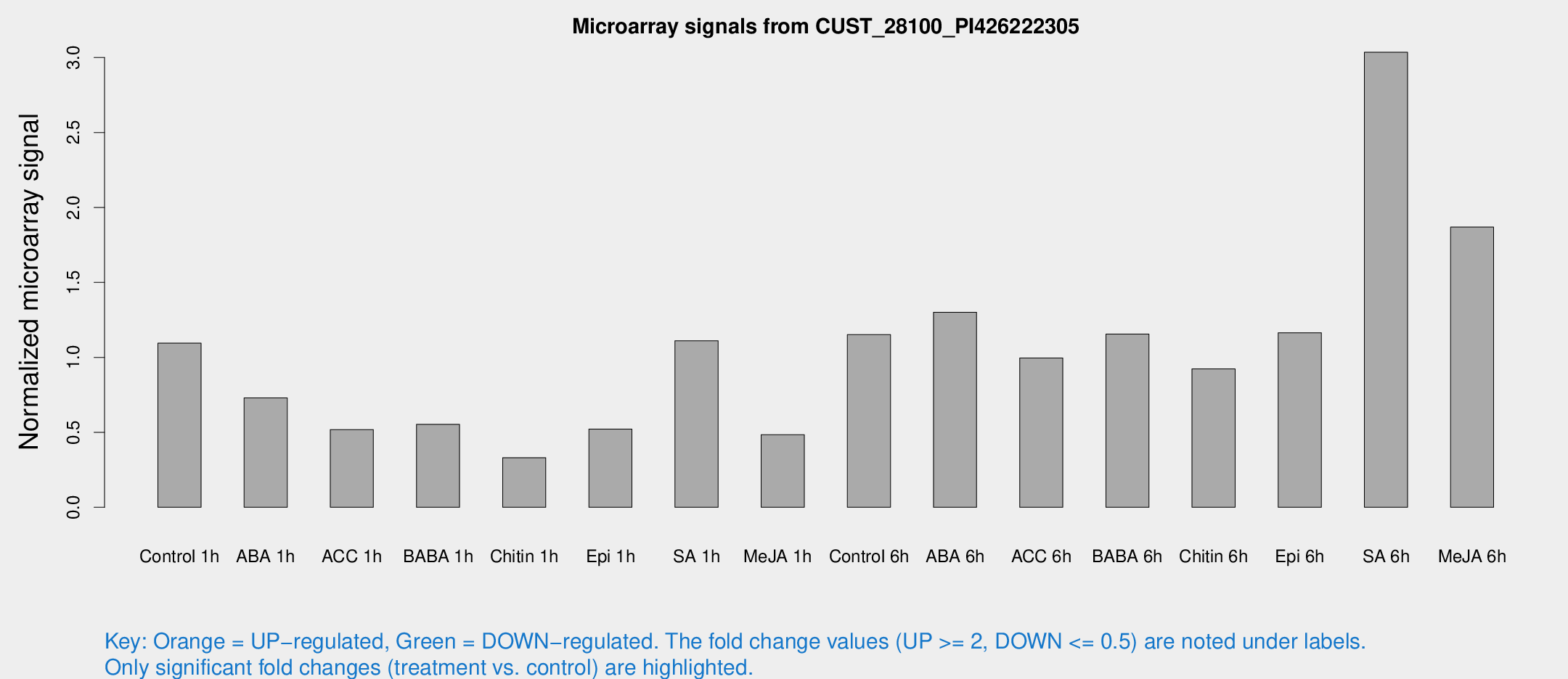
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1062.36 | 136.733 | 1.09479 | 0.13308 |
| ABA 1h | 731.991 | 289.874 | 0.729796 | 0.328913 |
| ACC 1h | 678.555 | 353.272 | 0.518543 | 0.305764 |
| BABA 1h | 584.707 | 183.729 | 0.55352 | 0.176403 |
| Chitin 1h | 321.139 | 117.62 | 0.331059 | 0.099865 |
| Epi 1h | 445.219 | 78.3104 | 0.521969 | 0.095043 |
| SA 1h | 1253.57 | 433.236 | 1.11047 | 0.399601 |
| Me-JA 1h | 425.097 | 153.387 | 0.483775 | 0.130401 |
| Control 6h | 1169.8 | 259.957 | 1.15138 | 0.197643 |
| ABA 6h | 1382.57 | 295.784 | 1.30123 | 0.214012 |
| ACC 6h | 1114.82 | 170.381 | 0.996181 | 0.212564 |
| BABA 6h | 1245.92 | 119.723 | 1.15549 | 0.0694524 |
| Chitin 6h | 955.089 | 126.487 | 0.923612 | 0.117594 |
| Epi 6h | 1279.32 | 175.221 | 1.16464 | 0.0974086 |
| SA 6h | 2997.02 | 581.99 | 3.03541 | 0.314938 |
| Me-JA 6h | 1890.2 | 495.518 | 1.86879 | 0.331345 |
Source Transcript PGSC0003DMT400004541 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G29420.1 | +1 | 2e-61 | 200 | 104/223 (47%) | glutathione S-transferase tau 7 | chr2:12618111-12618871 REVERSE LENGTH=227 |
