Probe CUST_28192_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28192_PI426222305 | JHI_St_60k_v1 | DMT400004540 | CAAAATTTGTGGATGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
All Microarray Probes Designed to Gene DMG400001804
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28106_PI426222305 | JHI_St_60k_v1 | DMT400004539 | ACTCGTGATTCTCGAGTATATCGATGAGACATGGAACGAAACAGCTCCTTTGCTACCTCA |
| CUST_28192_PI426222305 | JHI_St_60k_v1 | DMT400004540 | CAAAATTTGTGGATGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28178_PI426222305 | JHI_St_60k_v1 | DMT400004542 | AAGTCTCGTTTACCCCTTCAAATGCTCTCCTATTCCGCTTTTTTCAAATGATTGCTAAAG |
| CUST_28100_PI426222305 | JHI_St_60k_v1 | DMT400004541 | CAAAATTTGTGGACGACAAGCTTCTGCCATCAGTAACGAGTGTTTTCACAGAAAAAGGAT |
| CUST_28090_PI426222305 | JHI_St_60k_v1 | DMT400004537 | CTGCTTCATGAAAAATTTCAGACAGCAAATTAAATGGTGAAATTGCTGCAGTATGTGTCT |
Microarray Signals from CUST_28192_PI426222305
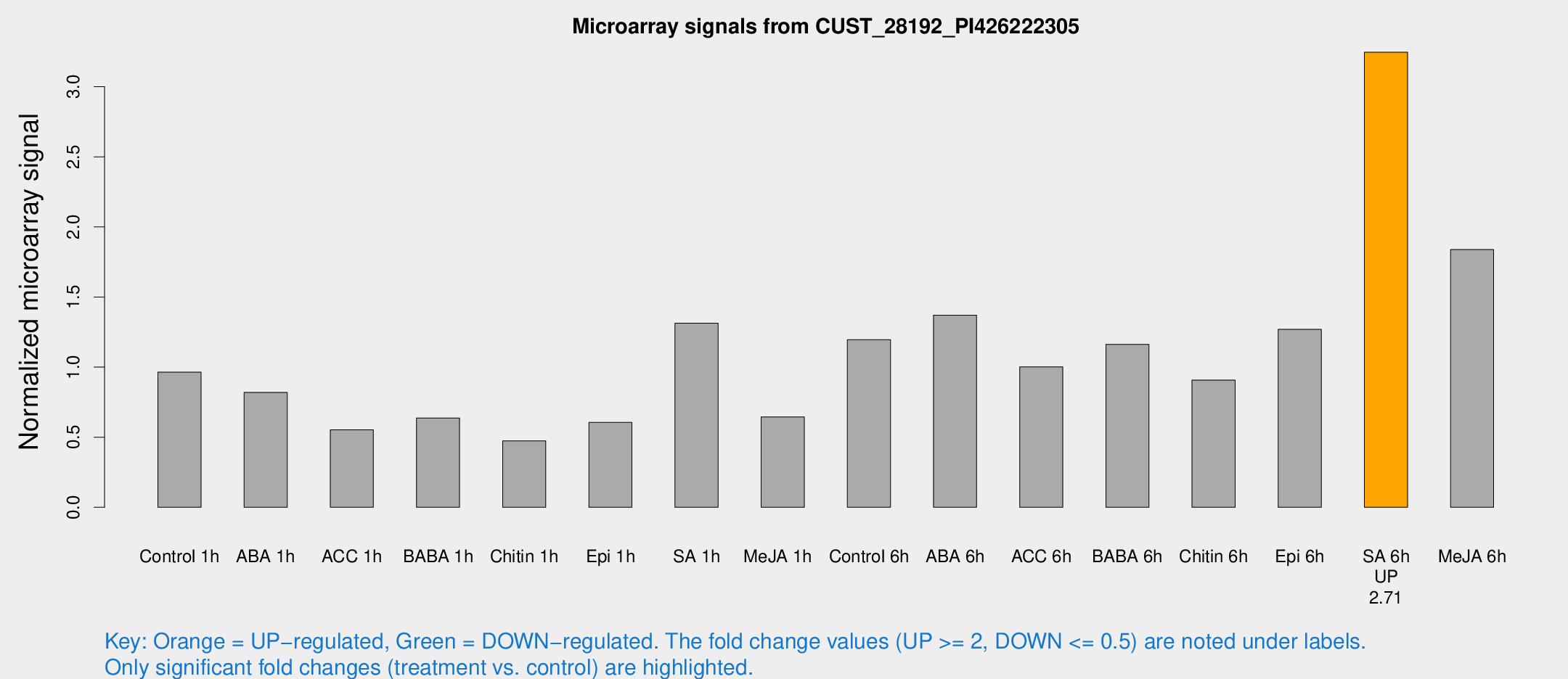
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1653.86 | 233.263 | 0.96384 | 0.0926674 |
| ABA 1h | 1330.05 | 366.207 | 0.819574 | 0.232102 |
| ACC 1h | 1151.39 | 444.087 | 0.553193 | 0.240053 |
| BABA 1h | 1149.36 | 329.106 | 0.637239 | 0.160475 |
| Chitin 1h | 755.221 | 177.846 | 0.474137 | 0.077831 |
| Epi 1h | 901.31 | 134.923 | 0.605617 | 0.0927069 |
| SA 1h | 2513.68 | 780.251 | 1.31365 | 0.357798 |
| Me-JA 1h | 940.723 | 234.059 | 0.645221 | 0.101409 |
| Control 6h | 2170.87 | 526.971 | 1.19622 | 0.239639 |
| ABA 6h | 2569.2 | 511.163 | 1.37078 | 0.238695 |
| ACC 6h | 1974.64 | 272.482 | 1.00207 | 0.174204 |
| BABA 6h | 2203.96 | 159.781 | 1.16271 | 0.067168 |
| Chitin 6h | 1665.06 | 246.495 | 0.907519 | 0.133626 |
| Epi 6h | 2432.81 | 229.248 | 1.2691 | 0.0733087 |
| SA 6h | 5590.39 | 949.256 | 3.24618 | 0.235178 |
| Me-JA 6h | 3225.38 | 707.371 | 1.839 | 0.248651 |
Source Transcript PGSC0003DMT400004540 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G29420.1 | +3 | 1e-61 | 199 | 105/224 (47%) | glutathione S-transferase tau 7 | chr2:12618111-12618871 REVERSE LENGTH=227 |
