Probe CUST_25432_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25432_PI426222305 | JHI_St_60k_v1 | DMT400034698 | AAAGAAATGGCCACTCTATCTTAGCACAAAAAATACCATCCTTAAGAAATACGACGGAAG |
All Microarray Probes Designed to Gene DMG400013332
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25375_PI426222305 | JHI_St_60k_v1 | DMT400034695 | AGACAGCGTGTCTAACGGCCTTTTGTTATAATCATTATAGTTCTTTCTTTGACTCTTCTT |
| CUST_25361_PI426222305 | JHI_St_60k_v1 | DMT400034696 | TAGCACTTATTATCCACGGATCCAAGATTTGGATCGCTTGGATTGATGACATCGATCCTG |
| CUST_25432_PI426222305 | JHI_St_60k_v1 | DMT400034698 | AAAGAAATGGCCACTCTATCTTAGCACAAAAAATACCATCCTTAAGAAATACGACGGAAG |
| CUST_25291_PI426222305 | JHI_St_60k_v1 | DMT400034697 | TTTCACTAAAAATGGCTTTTCAAAAGATCAAAGTGGCTAACCCCATAGTTGAAATGGACG |
| CUST_25460_PI426222305 | JHI_St_60k_v1 | DMT400034694 | TGAAAGCGCTGAGGCTACTTTGAAATGACCCGTGTCATTTGGAAATTAATTAAGGATAAG |
Microarray Signals from CUST_25432_PI426222305
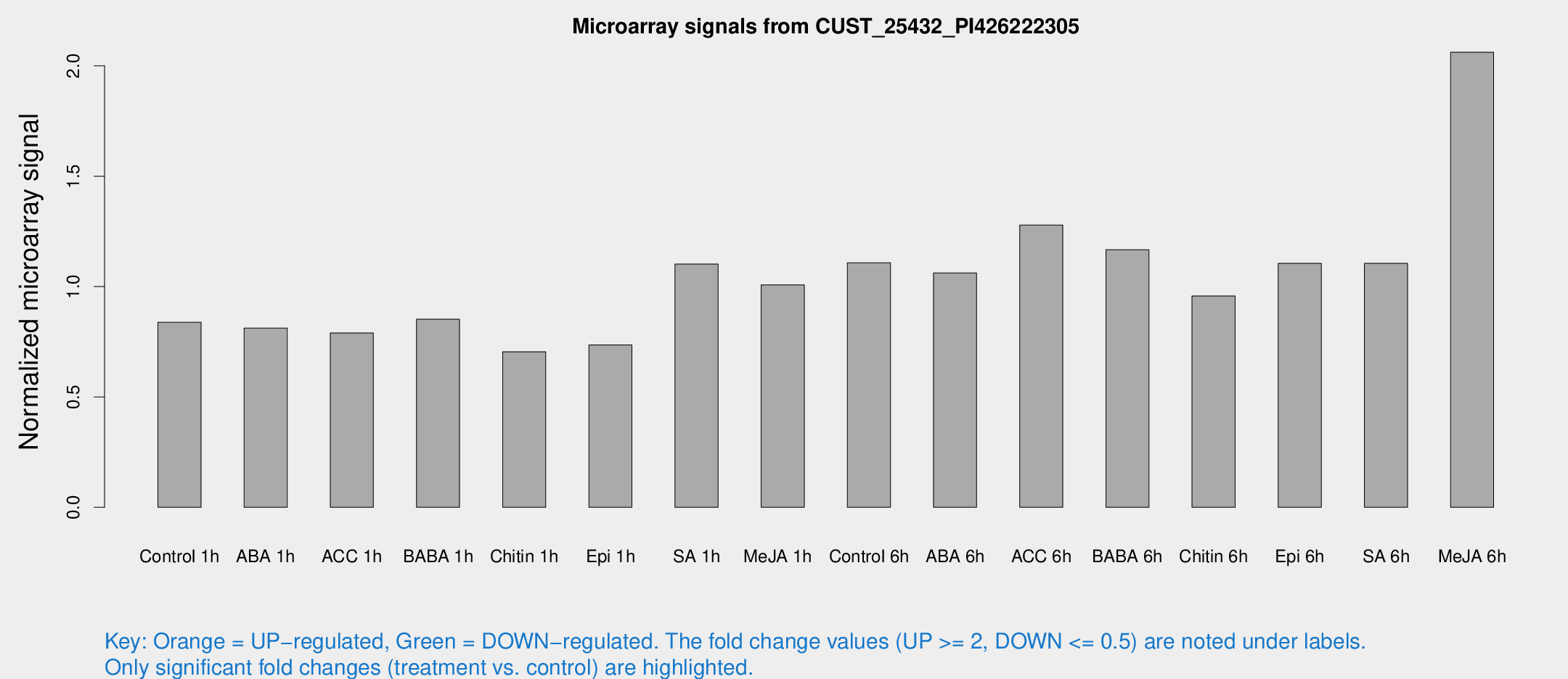
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 168.878 | 26.1591 | 0.838187 | 0.0967739 |
| ABA 1h | 142.822 | 14.6391 | 0.811763 | 0.0508475 |
| ACC 1h | 175.222 | 47.1384 | 0.789609 | 0.205069 |
| BABA 1h | 172.822 | 41.2062 | 0.852272 | 0.145104 |
| Chitin 1h | 128.114 | 21.6295 | 0.704664 | 0.0646352 |
| Epi 1h | 126.282 | 10.987 | 0.735621 | 0.0691189 |
| SA 1h | 227.76 | 32.3158 | 1.10182 | 0.128375 |
| Me-JA 1h | 164.115 | 18.6622 | 1.0077 | 0.0623496 |
| Control 6h | 254.616 | 80.929 | 1.10785 | 0.400832 |
| ABA 6h | 229.768 | 39.2824 | 1.06168 | 0.148255 |
| ACC 6h | 294.96 | 43.9155 | 1.2786 | 0.0761833 |
| BABA 6h | 258.793 | 19.8173 | 1.16726 | 0.0704786 |
| Chitin 6h | 200.764 | 12.318 | 0.957481 | 0.0587263 |
| Epi 6h | 251.191 | 39.8781 | 1.10502 | 0.16725 |
| SA 6h | 222.399 | 37.2123 | 1.10532 | 0.0739443 |
| Me-JA 6h | 412.461 | 57.8915 | 2.06185 | 0.120641 |
Source Transcript PGSC0003DMT400034698 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G65930.1 | +3 | 3e-139 | 406 | 193/222 (87%) | cytosolic NADP+-dependent isocitrate dehydrogenase | chr1:24539088-24541861 FORWARD LENGTH=410 |
