Probe CUST_23088_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23088_PI426222305 | JHI_St_60k_v1 | DMT400021129 | CCCATTTAGTGTAGGAGCCAGAATTTTCATTTTCTCCTGTAGGTAACATCTAAAGAAGTA |
All Microarray Probes Designed to Gene DMG400008183
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22942_PI426222305 | JHI_St_60k_v1 | DMT400021130 | GCGACTTCCAATCCACTAGTCAATCAGAAATCAACAAAGAGAAAGGAACGCGAGAAAAAG |
| CUST_23014_PI426222305 | JHI_St_60k_v1 | DMT400021132 | ATTAGCGAAGTGGTTCCCATGTGGAGAGTAACTTACAAGACATGGTTGTGCCACCTATCT |
| CUST_23088_PI426222305 | JHI_St_60k_v1 | DMT400021129 | CCCATTTAGTGTAGGAGCCAGAATTTTCATTTTCTCCTGTAGGTAACATCTAAAGAAGTA |
Microarray Signals from CUST_23088_PI426222305
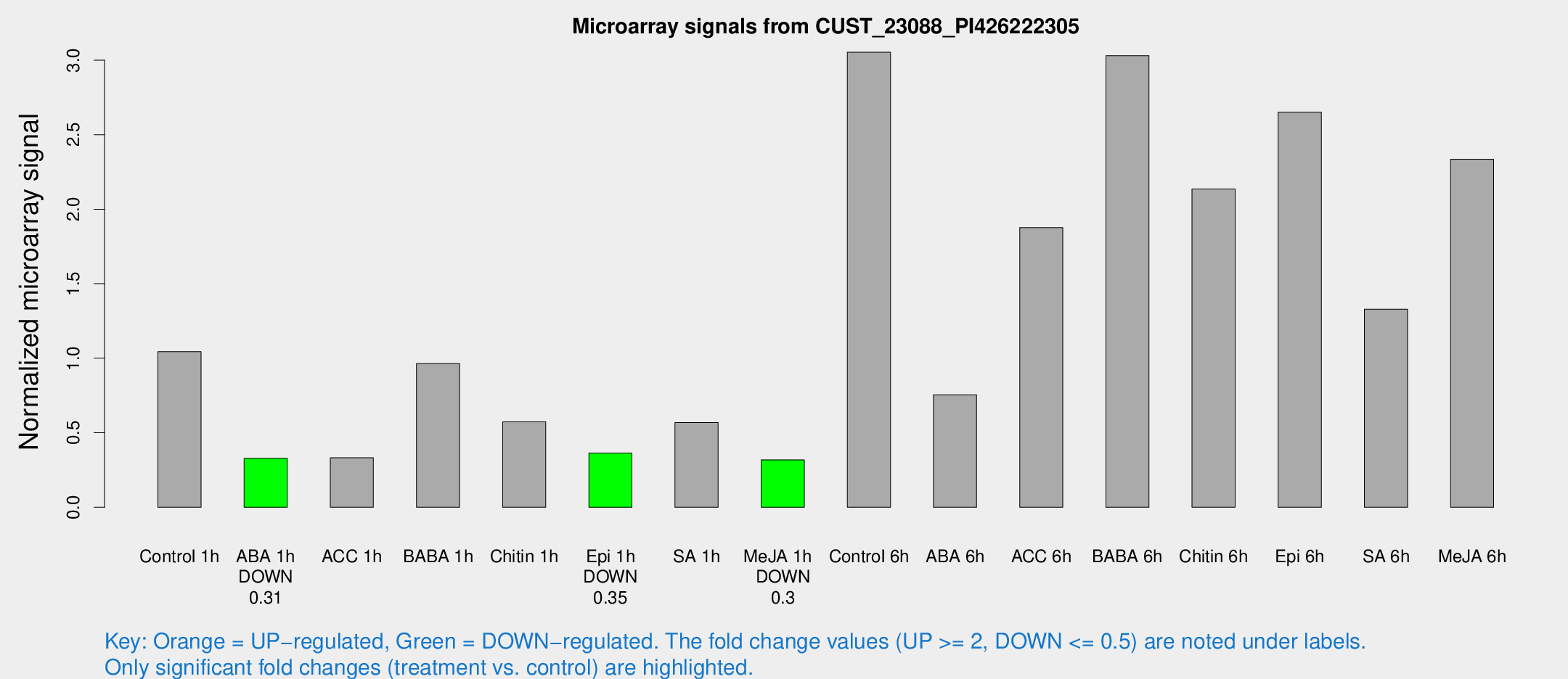
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.9456 | 3.25918 | 1.04453 | 0.157819 |
| ABA 1h | 6.09003 | 2.92953 | 0.328904 | 0.160556 |
| ACC 1h | 7.60094 | 3.33451 | 0.332176 | 0.163629 |
| BABA 1h | 19.9625 | 3.39084 | 0.964601 | 0.171735 |
| Chitin 1h | 13.5728 | 6.69281 | 0.5737 | 0.27366 |
| Epi 1h | 6.83526 | 3.00495 | 0.364242 | 0.174328 |
| SA 1h | 13.3909 | 3.77729 | 0.568862 | 0.230301 |
| Me-JA 1h | 5.41054 | 3.10728 | 0.318364 | 0.181134 |
| Control 6h | 76.7045 | 26.3852 | 3.05312 | 1.29313 |
| ABA 6h | 16.8194 | 3.40537 | 0.755158 | 0.154004 |
| ACC 6h | 48.441 | 12.4788 | 1.87605 | 0.360956 |
| BABA 6h | 77.1573 | 20.1096 | 3.03024 | 0.894444 |
| Chitin 6h | 53.9514 | 18.2791 | 2.13615 | 0.923155 |
| Epi 6h | 74.476 | 32.0193 | 2.65136 | 1.34373 |
| SA 6h | 36.4167 | 16.3922 | 1.32932 | 1.32727 |
| Me-JA 6h | 57.8005 | 20.7557 | 2.3355 | 0.954936 |
Source Transcript PGSC0003DMT400021129 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
