Probe CUST_16496_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16496_PI426222305 | JHI_St_60k_v1 | DMT400069306 | GTGCATGCTACTATGATGAACAACCTAATAATGTGAAAGTGAAATCTCTTGTGGTATGAA |
All Microarray Probes Designed to Gene DMG400026959
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16802_PI426222305 | JHI_St_60k_v1 | DMT400069305 | TCTCATCATGACCAAAATATTCATCTCAAGTATAGTACTTGCCCAACATCACTACACTAG |
| CUST_16496_PI426222305 | JHI_St_60k_v1 | DMT400069306 | GTGCATGCTACTATGATGAACAACCTAATAATGTGAAAGTGAAATCTCTTGTGGTATGAA |
Microarray Signals from CUST_16496_PI426222305
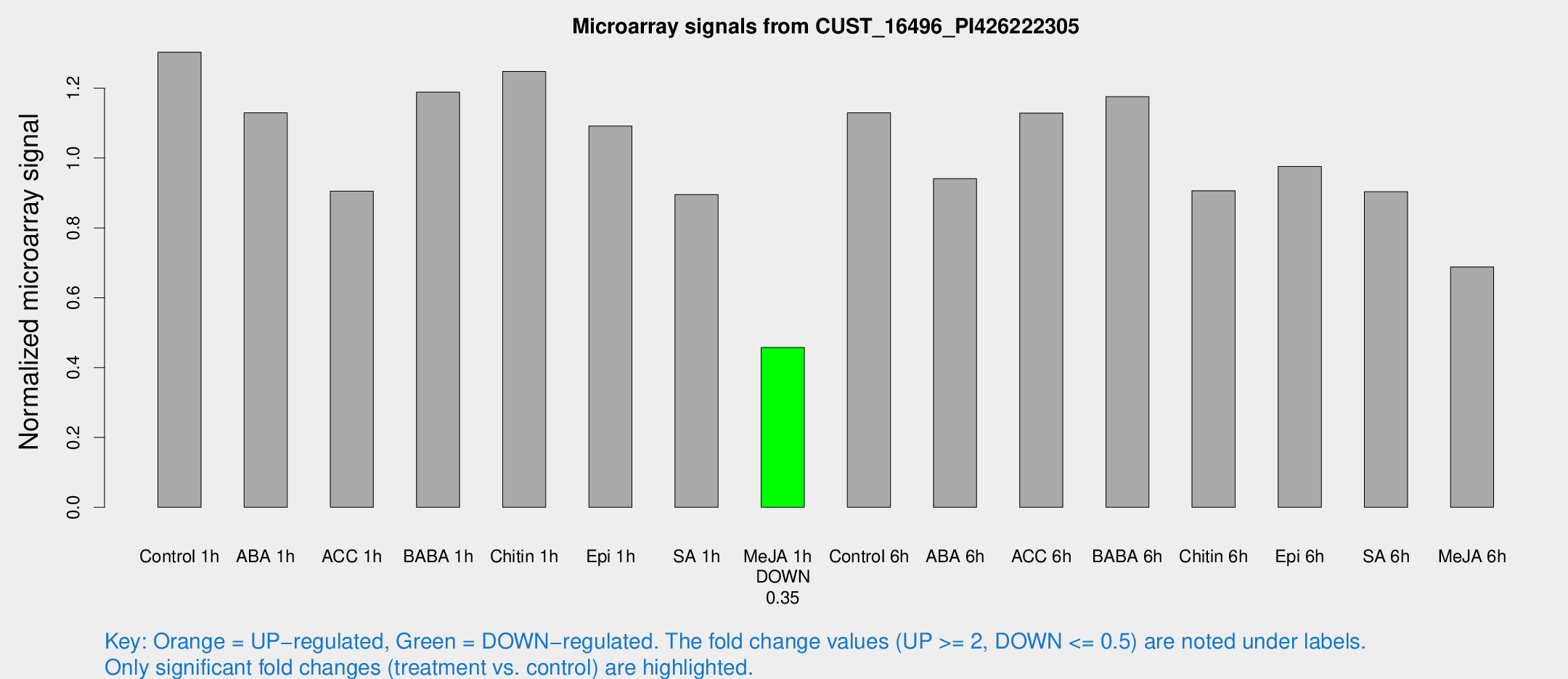
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 351.181 | 44.254 | 1.30297 | 0.0901567 |
| ABA 1h | 265.321 | 15.7359 | 1.12949 | 0.0819256 |
| ACC 1h | 250.516 | 29.4115 | 0.905264 | 0.0547144 |
| BABA 1h | 307.131 | 28.9015 | 1.18868 | 0.0703997 |
| Chitin 1h | 301.242 | 32.5334 | 1.24772 | 0.077044 |
| Epi 1h | 262.679 | 52.7774 | 1.09147 | 0.22571 |
| SA 1h | 245.794 | 18.9542 | 0.895373 | 0.090153 |
| Me-JA 1h | 99.2253 | 6.84093 | 0.457638 | 0.0351493 |
| Control 6h | 344.159 | 109.866 | 1.12944 | 0.371408 |
| ABA 6h | 278.317 | 59.2949 | 0.9413 | 0.243247 |
| ACC 6h | 363.865 | 77.8663 | 1.12862 | 0.217933 |
| BABA 6h | 351.393 | 24.8106 | 1.17593 | 0.103553 |
| Chitin 6h | 257.409 | 17.6127 | 0.906523 | 0.0690754 |
| Epi 6h | 295.005 | 28.4713 | 0.975712 | 0.0582659 |
| SA 6h | 240.14 | 24.9769 | 0.903593 | 0.0545126 |
| Me-JA 6h | 188.151 | 35.4591 | 0.688107 | 0.118587 |
Source Transcript PGSC0003DMT400069306 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G01940.3 | +1 | 2e-131 | 394 | 224/402 (56%) | C2H2-like zinc finger protein | chr2:432652-434917 FORWARD LENGTH=446 |
