Probe CUST_12890_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12890_PI426222305 | JHI_St_60k_v1 | DMT400063326 | GATCAGCCTAATTCGAATTTGTCTTTAGAGCTTGGAGAAGGAAGAGATAAGATGCCCCTA |
All Microarray Probes Designed to Gene DMG400024632
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12949_PI426222305 | JHI_St_60k_v1 | DMT400063327 | CTGGATTATTCATGAGCAAACATAATCATGCTACATTATTCCTTCTTCACAGGTACAACA |
| CUST_12890_PI426222305 | JHI_St_60k_v1 | DMT400063326 | GATCAGCCTAATTCGAATTTGTCTTTAGAGCTTGGAGAAGGAAGAGATAAGATGCCCCTA |
Microarray Signals from CUST_12890_PI426222305
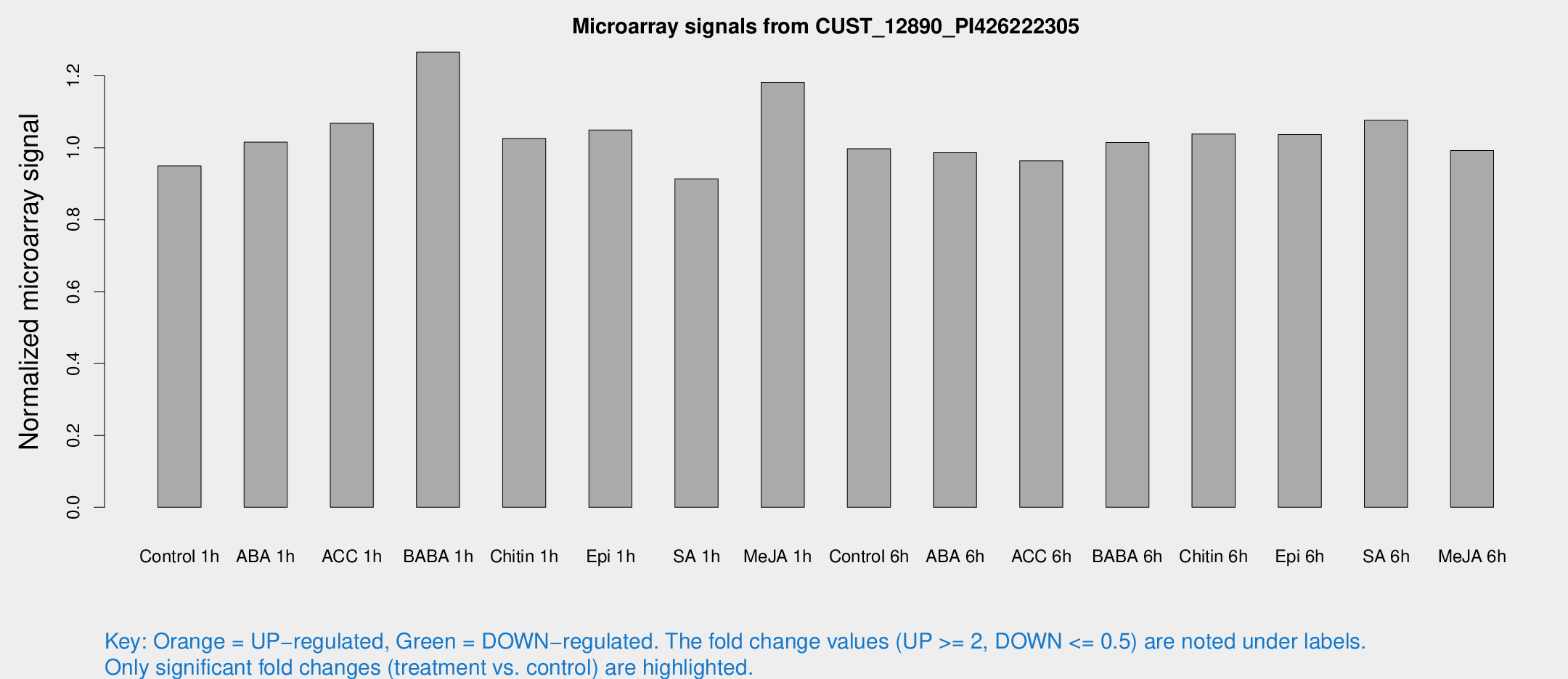
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.41028 | 3.71804 | 0.949466 | 0.549791 |
| ABA 1h | 6.05575 | 3.50741 | 1.01563 | 0.588203 |
| ACC 1h | 7.51079 | 4.42868 | 1.06767 | 0.618712 |
| BABA 1h | 8.5123 | 3.8921 | 1.26559 | 0.625405 |
| Chitin 1h | 6.25257 | 3.64174 | 1.02587 | 0.594158 |
| Epi 1h | 6.15451 | 3.58507 | 1.04922 | 0.607976 |
| SA 1h | 6.32726 | 3.66959 | 0.912953 | 0.528932 |
| Me-JA 1h | 6.53652 | 3.80745 | 1.18199 | 0.685013 |
| Control 6h | 6.74501 | 3.9127 | 0.99722 | 0.577799 |
| ABA 6h | 7.06707 | 4.02698 | 0.985984 | 0.561424 |
| ACC 6h | 7.59555 | 4.49738 | 0.963562 | 0.558995 |
| BABA 6h | 7.67898 | 4.29599 | 1.01436 | 0.565961 |
| Chitin 6h | 7.45463 | 4.32526 | 1.03796 | 0.601602 |
| Epi 6h | 8.05598 | 4.77411 | 1.03683 | 0.600382 |
| SA 6h | 7.18157 | 4.16221 | 1.07658 | 0.623609 |
| Me-JA 6h | 6.66033 | 3.85718 | 0.99222 | 0.574569 |
Source Transcript PGSC0003DMT400063326 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G02150.2 | +1 | 3e-43 | 151 | 80/147 (54%) | plastid transcription factor 1 | chr3:391522-392589 FORWARD LENGTH=355 |
