Probe CUST_11038_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11038_PI426222305 | JHI_St_60k_v1 | DMT400078541 | ACGAGAAGCTACTGATATATGACTTCATTCCAAATGGAAACCTTACCACCGCAATTCACG |
All Microarray Probes Designed to Gene DMG400030572
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11038_PI426222305 | JHI_St_60k_v1 | DMT400078541 | ACGAGAAGCTACTGATATATGACTTCATTCCAAATGGAAACCTTACCACCGCAATTCACG |
| CUST_11308_PI426222305 | JHI_St_60k_v1 | DMT400078542 | ACGAGAAGCTACTGATATATGACTTCATTCCAAATGGAAACCTTACCACCGCAATTCACG |
Microarray Signals from CUST_11038_PI426222305
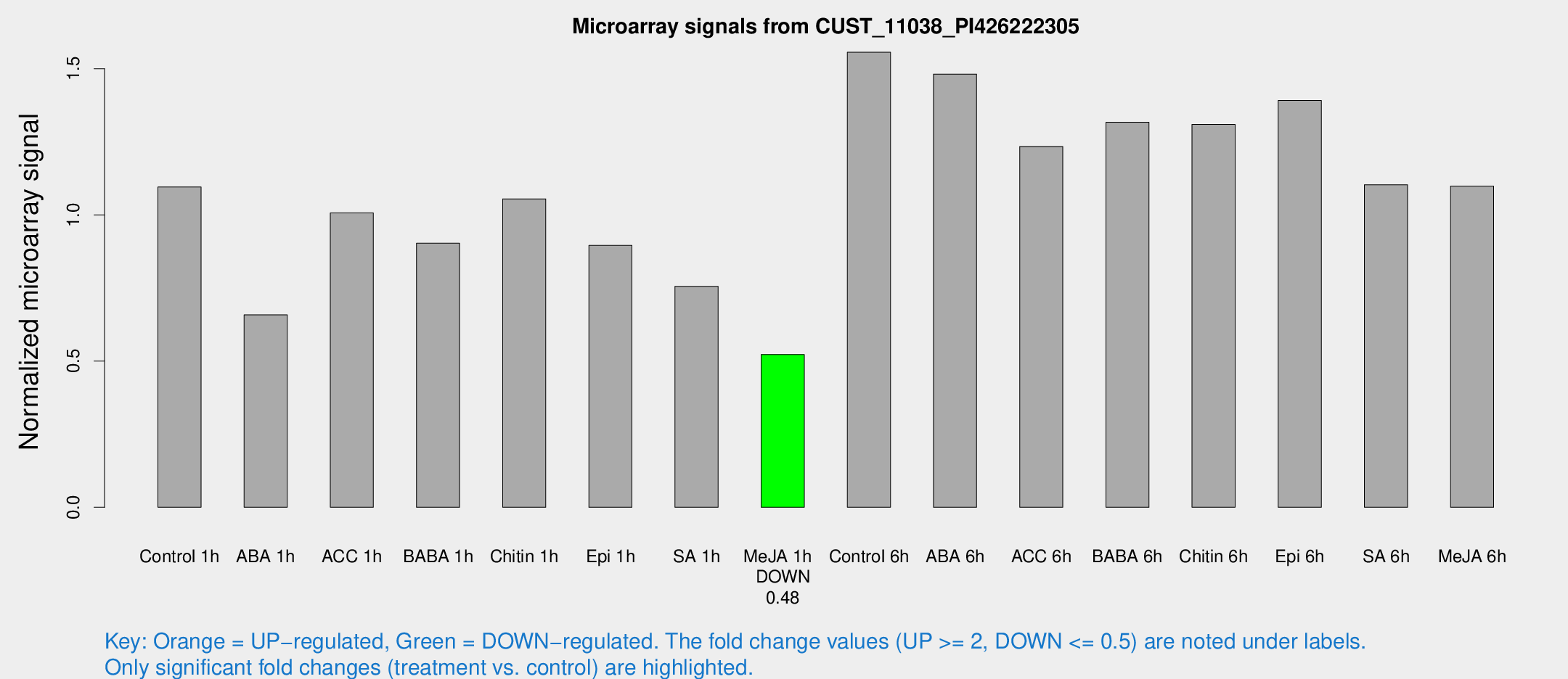
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 137.554 | 36.6217 | 1.0957 | 0.211522 |
| ABA 1h | 68.8936 | 5.14083 | 0.658302 | 0.0490467 |
| ACC 1h | 135.947 | 42.4655 | 1.00708 | 0.283685 |
| BABA 1h | 103.75 | 10.9671 | 0.903169 | 0.0626337 |
| Chitin 1h | 111.854 | 7.44387 | 1.0549 | 0.0865002 |
| Epi 1h | 91.7961 | 7.02514 | 0.895736 | 0.0626736 |
| SA 1h | 92.5562 | 10.044 | 0.755587 | 0.0581781 |
| Me-JA 1h | 51.6674 | 8.37534 | 0.522504 | 0.0706084 |
| Control 6h | 202.484 | 54.8702 | 1.5565 | 0.393047 |
| ABA 6h | 186.205 | 11.767 | 1.48165 | 0.0916105 |
| ACC 6h | 179.635 | 51.6899 | 1.23422 | 0.163636 |
| BABA 6h | 180.055 | 31.9562 | 1.31692 | 0.218826 |
| Chitin 6h | 165.901 | 17.0224 | 1.30972 | 0.119073 |
| Epi 6h | 196.622 | 51.0651 | 1.39176 | 0.382968 |
| SA 6h | 130.747 | 17.7377 | 1.10338 | 0.123282 |
| Me-JA 6h | 143.624 | 42.2695 | 1.09853 | 0.306241 |
Source Transcript PGSC0003DMT400078541 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G01210.1 | +1 | 0.0 | 607 | 326/499 (65%) | Leucine-rich repeat protein kinase family protein | chr2:119509-121734 REVERSE LENGTH=716 |
