Probe CUST_10702_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10702_PI426222305 | JHI_St_60k_v1 | DMT400031582 | AAACTGCCAACTATGCCTTAAAATTGGGAATTGATCAAGGGAAAAGCTATGTGAAGTAAA |
All Microarray Probes Designed to Gene DMG400012117
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10702_PI426222305 | JHI_St_60k_v1 | DMT400031582 | AAACTGCCAACTATGCCTTAAAATTGGGAATTGATCAAGGGAAAAGCTATGTGAAGTAAA |
| CUST_10867_PI426222305 | JHI_St_60k_v1 | DMT400031580 | GCAGTAAAATATCCTGGATCAGTCAAATCCCTCACTTTAATTGCACCGGTTTCCGAAGAA |
| CUST_10634_PI426222305 | JHI_St_60k_v1 | DMT400031581 | GCAAGGGAGCTTGAACAAATTTGGAATAATTCAACATGTTCAAAATAATTTCCCACCATC |
Microarray Signals from CUST_10702_PI426222305
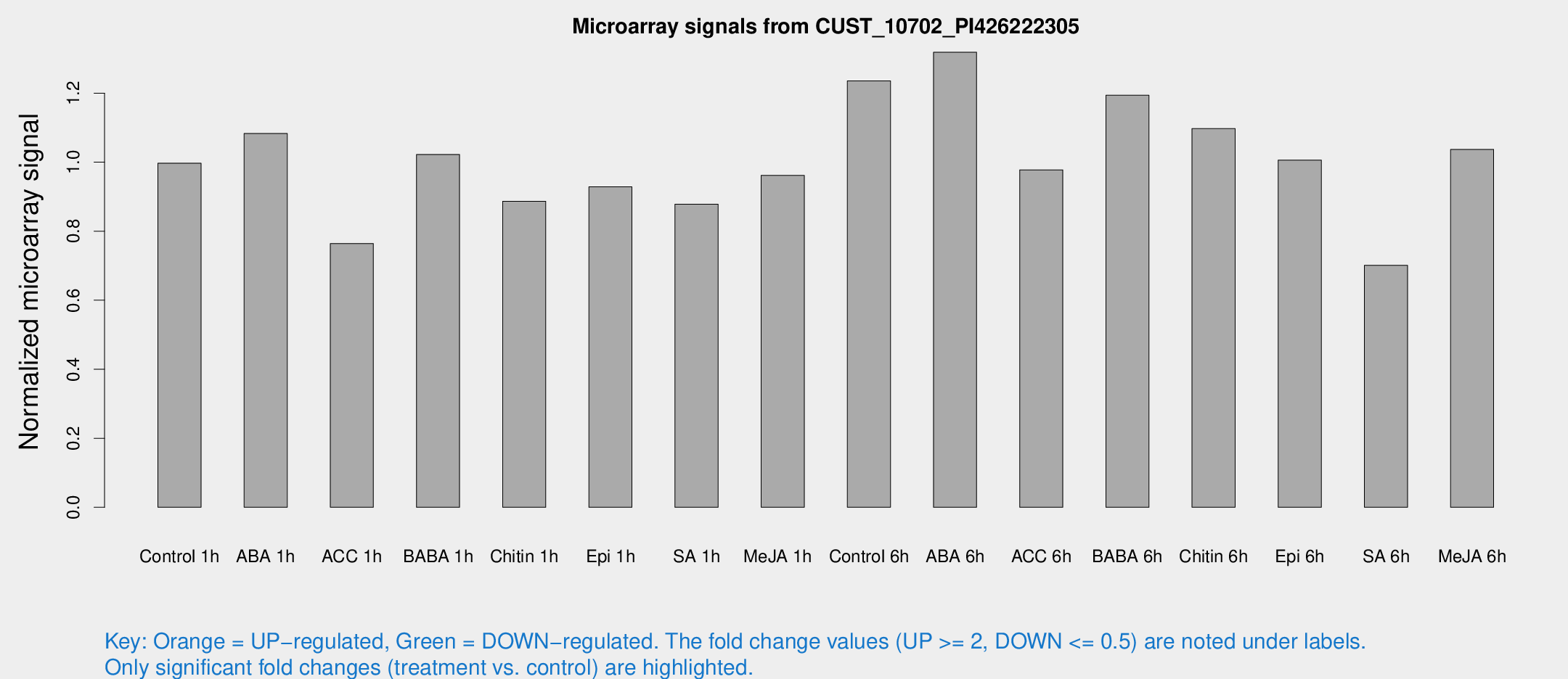
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1714.85 | 165.449 | 0.99697 | 0.0575974 |
| ABA 1h | 1643.66 | 130.134 | 1.08288 | 0.076676 |
| ACC 1h | 1479.52 | 421.827 | 0.7643 | 0.21025 |
| BABA 1h | 1691.88 | 143.633 | 1.02222 | 0.0624571 |
| Chitin 1h | 1377.04 | 149.208 | 0.88678 | 0.0559494 |
| Epi 1h | 1375.06 | 80.8088 | 0.928503 | 0.0536621 |
| SA 1h | 1556.39 | 170.971 | 0.878389 | 0.0933807 |
| Me-JA 1h | 1352.17 | 133.202 | 0.961593 | 0.0555797 |
| Control 6h | 2205.77 | 427.235 | 1.23549 | 0.162445 |
| ABA 6h | 2492.42 | 495.773 | 1.31852 | 0.205449 |
| ACC 6h | 1969.72 | 334.992 | 0.97766 | 0.0701362 |
| BABA 6h | 2443.02 | 577.589 | 1.19392 | 0.296587 |
| Chitin 6h | 2037.99 | 294.43 | 1.09711 | 0.18344 |
| Epi 6h | 2039.72 | 462.227 | 1.00613 | 0.2913 |
| SA 6h | 1773.62 | 787.497 | 0.701242 | 1.07305 |
| Me-JA 6h | 1899.42 | 507.748 | 1.03687 | 0.207779 |
Source Transcript PGSC0003DMT400031582 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
