- Transcript BART1_0-u42531.005
- Transcript IDBART1_0-u42531.005
- Gene IDBART1_0-u42531
Exon Structure of BART1_0-u42531.005
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr6H | 1 | 22236688 | 22236580 | + |
| chr6H | 2 | 22237825 | 22237685 | + |
| chr6H | 3 | 22239292 | 22239057 | + |
| chr6H | 4 | 22239749 | 22239402 | + |
| chr6H | 5 | 22240129 | 22239832 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials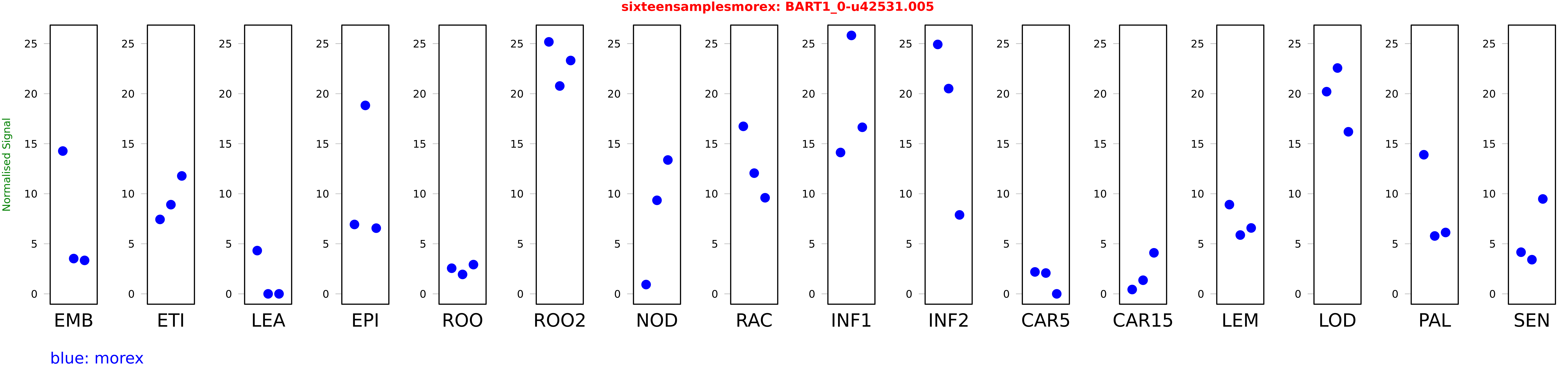
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 14.2714 | 3.52583 | 3.34511 |
| morex | ETI | 7.43901 | 8.91218 | 11.7839 |
| morex | LEA | 4.32265 | 0 | 0.00000182611 |
| morex | EPI | 6.9341 | 18.8312 | 6.56538 |
| morex | ROO | 2.56149 | 1.94174 | 2.92235 |
| morex | ROO2 | 25.1791 | 20.7709 | 23.3185 |
| morex | NOD | 0.929963 | 9.34779 | 13.3755 |
| morex | RAC | 16.7393 | 12.0621 | 9.60244 |
| morex | INF1 | 14.1255 | 25.8239 | 16.6451 |
| morex | INF2 | 24.9236 | 20.5116 | 7.88925 |
| morex | CAR5 | 2.19107 | 2.08058 | 0 |
| morex | CAR15 | 0.433645 | 1.36213 | 4.09988 |
| morex | LEM | 8.91658 | 5.87738 | 6.58529 |
| morex | LOD | 20.2041 | 22.5679 | 16.1957 |
| morex | PAL | 13.9012 | 5.78309 | 6.13334 |
| morex | SEN | 4.16278 | 3.4144 | 9.48229 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os02g03040.4 | +2 | 9e-78 | 240 | 144/171 (84%) | protein|RNA recognition motif containing protein, expressed |
| TAIR PP10 | AT5G52040 @ ARAPORT AT5G52040.4 @ TAIR |
+2 | 3e-46 | 161 | 103/194 (53%) | Symbols: RS41, At-RS41 | RNA-binding (RRM/RBD/RNP motifs) family protein | chr5:21131881-21133318 FORWARD LENGTH=324 |
| BRACH PP3 | Bradi3g02226.1.p | +2 | 2e-88 | 269 | 158/168 (94%) | pacid=32814082 transcript=Bradi3g02226.1 locus=Bradi3g02226 ID=Bradi3g02226.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (1132 bp)
>BART1_0-u42531.005 1132 150831_barley_pseudomolecules TGCCCCTGTCTGTCTCCGTCCTCCCCCCGCTCCACTCGCCGCCGGGTCCGAACCAGCCGC GCCCCCCGTCGAAAACCTAGGGTTCCGCCCTCCCCCGCCCCCTCCGAAGGTTTGGGGAAG ATCTGTTTATACAGCAACACCAACATGAGGCCAATTTTCTGCGGGAACCTTGACTATGAT GCCCGCCAATCTGAAGTCGAGCGTCTTTTTGGCAAATATGGAAGAGTGGAGCGAGTTGAC ATGAAGACAGGAGGATCGCAGTGGTGGTCGGAAAGGAAATGGAAAGAGATCTCCAAGCAG CGTGAAACCAACCAAAACCTTGTTTGTGATCAACTTTGATCCAATCAACACAAGGACAAG AGATCTGGAGAAGCACTTTGATCTGTATGGCAAGATTGCAAATATCAGAATCAGAAGGAA CTTTGCATTTGTCCAGTATGAAACACAGGAGGATGCTACTAAAGCTCTGGACGGTACTAA TGGAAGCACTGTGATGGACAGGGTGATTTCTGTTGAGTATGCGCTTCGTGACGATGATGA AAAAAGGAACGGGTACAGTCCAGACAGGAGAGGAGGCCGTGATAGGTCTCCTGACAGAAG GGACAATCGTGGTAGGTCGGCAAGTCCTTATGGCAGAGGGCGGGAAAGGGGCAGCCCAGA CTATGGCAGGGGCAGGGAGAGGGGCAGCCCGGACTATGGCAAGGGCGGTGCCAGAGATAG TCCTGACTATGTCCGTGGTGGAAGCCCGTACGGCGGCAAAGGGGATGACAGGGCTAGCCC CAAGTATGACCGTGAACGCCGTGAAGCCAGTCCTGCCTATGACAGGCGCCGCAGCCGCTC GCCTGCAAGGGAAGATAGGGACTGAAGCTGTACCATGGATGGCTGCGCTCAATCCTGGGT GCTTCGGACCAAATGGATGCACCGTCCTAGTGCGATGTTCGCTCAAGTTGGTTATGTTAG GCAGCCAGGAACTTCTGTAATATCTCTCTATTAGCTAGCTCTAGCTGAATGCTTTGTCGA TTTAGTTTGAACTGTGATTGTTAGCTTGAACTTTGTGCAAAGGGATTCTTCCGGTGAGAC CATTTGCGGAGCGATGAGATGTACCGTGTATCGACCGACAGCAGTATTCTAG
Protein Sequence (123 aa)
>BART1_0-u42531.005 123 150831_barley_pseudomolecules MDRVISVEYALRDDDEKRNGYSPDRRGGRDRSPDRRDNRGRSASPYGRGRERGSPDYGRG RERGSPDYGKGGARDSPDYVRGGSPYGGKGDDRASPKYDRERREASPAYDRRRSRSPARE DRD
