Probe CUST_8583_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8583_PI426222305 | JHI_St_60k_v1 | DMT400058381 | GTGCCCTACGTTACACTCCATAGGATATTTAAGATTGTTTATACTTTTCACTTGAAGTAC |
All Microarray Probes Designed to Gene DMG400022678
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8583_PI426222305 | JHI_St_60k_v1 | DMT400058381 | GTGCCCTACGTTACACTCCATAGGATATTTAAGATTGTTTATACTTTTCACTTGAAGTAC |
Microarray Signals from CUST_8583_PI426222305
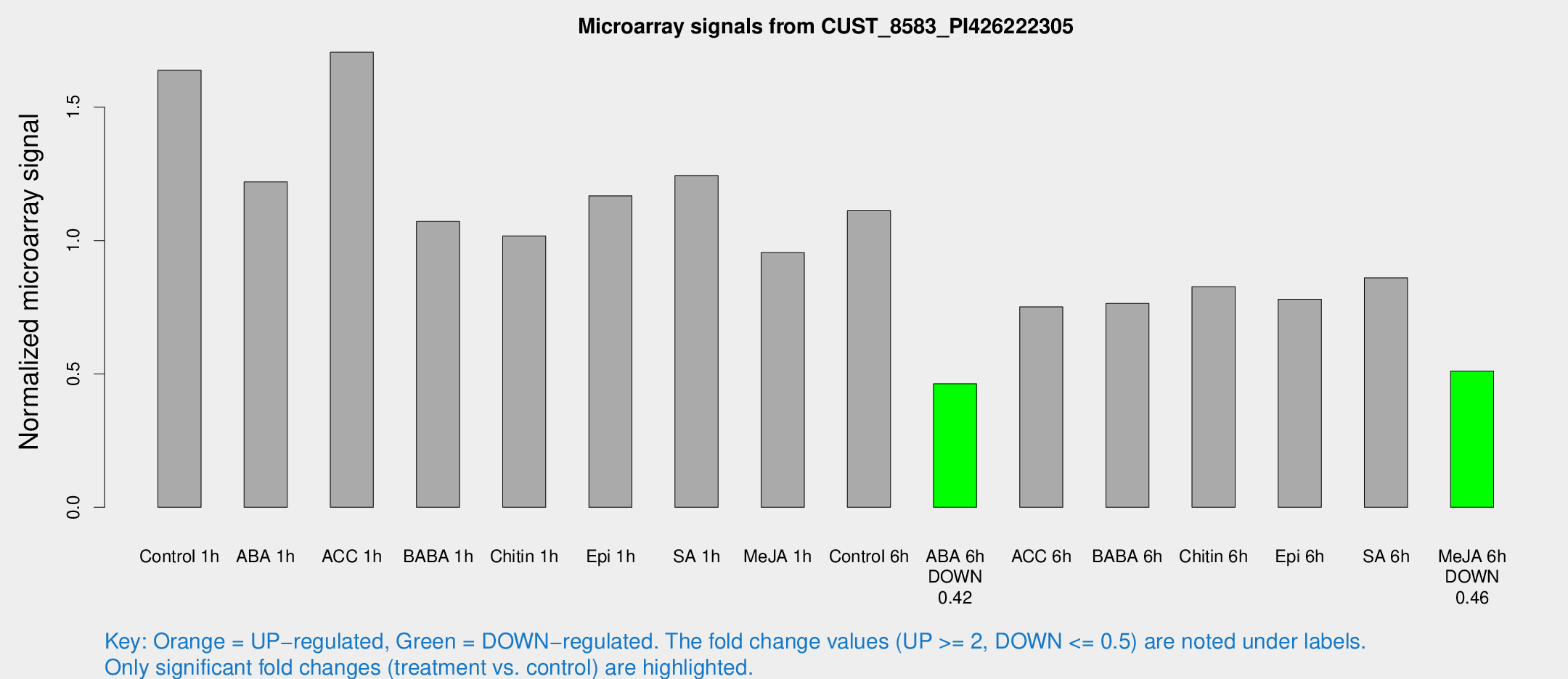
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1137.24 | 125.093 | 1.63826 | 0.094707 |
| ABA 1h | 749.523 | 82.0318 | 1.22014 | 0.173072 |
| ACC 1h | 1208.71 | 98.8242 | 1.70614 | 0.109101 |
| BABA 1h | 716.876 | 77.6537 | 1.07155 | 0.0620992 |
| Chitin 1h | 650.964 | 131.397 | 1.01728 | 0.205787 |
| Epi 1h | 694.649 | 40.333 | 1.16789 | 0.0676393 |
| SA 1h | 885.606 | 90.2655 | 1.24387 | 0.090983 |
| Me-JA 1h | 535.198 | 31.1275 | 0.95505 | 0.102131 |
| Control 6h | 792.068 | 139.483 | 1.11182 | 0.119291 |
| ABA 6h | 339.013 | 20.9958 | 0.463343 | 0.0271731 |
| ACC 6h | 606.197 | 97.5445 | 0.75121 | 0.0436523 |
| BABA 6h | 617.168 | 134.476 | 0.764337 | 0.199765 |
| Chitin 6h | 610.529 | 66.6464 | 0.82723 | 0.0623045 |
| Epi 6h | 608.911 | 59.3468 | 0.779896 | 0.139496 |
| SA 6h | 593.623 | 75.2376 | 0.860121 | 0.049973 |
| Me-JA 6h | 363.734 | 70.0353 | 0.510678 | 0.0670205 |
Source Transcript PGSC0003DMT400058381 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18660.1 | +2 | 0.0 | 593 | 301/394 (76%) | NAD(P)-binding Rossmann-fold superfamily protein | chr5:6220872-6222125 REVERSE LENGTH=417 |
