Probe CUST_8143_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8143_PI426222305 | JHI_St_60k_v1 | DMT400075617 | CGTGTTCTTGAAAAAGGATTGCTTCATAATCCATATGAATCGTGACGTTAATTCGATATG |
All Microarray Probes Designed to Gene DMG400029412
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8293_PI426222305 | JHI_St_60k_v1 | DMT400075618 | CGTGTTCTTGAAAAAGGATTGCTTCATAATCCATATGAATCGTGACGTTAATTCGATATG |
| CUST_7999_PI426222305 | JHI_St_60k_v1 | DMT400075616 | TTTGCATCTTTACTAGTCCCTACTTTTTCGTGTTGCTTTAGGAATTGGAACTAGACAAGT |
| CUST_8143_PI426222305 | JHI_St_60k_v1 | DMT400075617 | CGTGTTCTTGAAAAAGGATTGCTTCATAATCCATATGAATCGTGACGTTAATTCGATATG |
Microarray Signals from CUST_8143_PI426222305
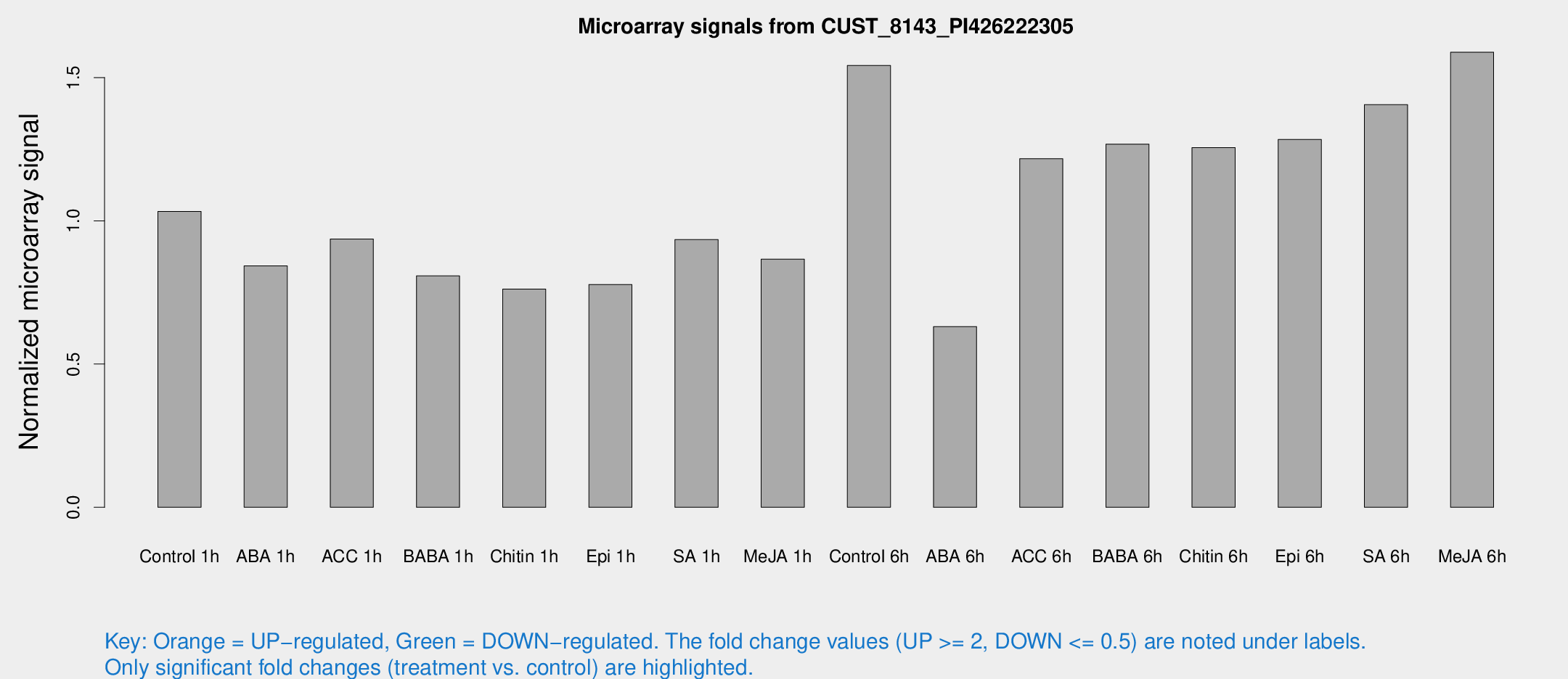
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1045.94 | 134.843 | 1.03289 | 0.0638097 |
| ABA 1h | 755.403 | 101.839 | 0.842728 | 0.0728822 |
| ACC 1h | 977.542 | 131.206 | 0.936583 | 0.0616422 |
| BABA 1h | 815.123 | 166.682 | 0.807768 | 0.104532 |
| Chitin 1h | 693.143 | 87.035 | 0.761443 | 0.0817798 |
| Epi 1h | 672.645 | 39.1166 | 0.777619 | 0.0450813 |
| SA 1h | 982.589 | 158.524 | 0.934237 | 0.130556 |
| Me-JA 1h | 708.741 | 53.7657 | 0.866361 | 0.0502266 |
| Control 6h | 1615.45 | 321.184 | 1.54233 | 0.208463 |
| ABA 6h | 673.946 | 62.7441 | 0.630791 | 0.0536684 |
| ACC 6h | 1429.17 | 242.15 | 1.21677 | 0.0703479 |
| BABA 6h | 1436.69 | 189.329 | 1.26774 | 0.155233 |
| Chitin 6h | 1337.52 | 91.7072 | 1.25573 | 0.0726089 |
| Epi 6h | 1457.97 | 153.129 | 1.28402 | 0.194662 |
| SA 6h | 1444.63 | 271.86 | 1.4055 | 0.134249 |
| Me-JA 6h | 1641.24 | 329.749 | 1.58845 | 0.189972 |
Source Transcript PGSC0003DMT400075617 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G42250.1 | +2 | 4e-96 | 307 | 143/203 (70%) | Zinc-binding alcohol dehydrogenase family protein | chr5:16894087-16897450 FORWARD LENGTH=390 |
