Probe CUST_802_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_802_PI426222305 | JHI_St_60k_v1 | DMT400003459 | TGTAGATATCGTCGAAAGTGAATGAGTTTAGTTGAATTCAAAGGCGGACCGCGGACCTAC |
All Microarray Probes Designed to Gene DMG400001366
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_802_PI426222305 | JHI_St_60k_v1 | DMT400003459 | TGTAGATATCGTCGAAAGTGAATGAGTTTAGTTGAATTCAAAGGCGGACCGCGGACCTAC |
Microarray Signals from CUST_802_PI426222305
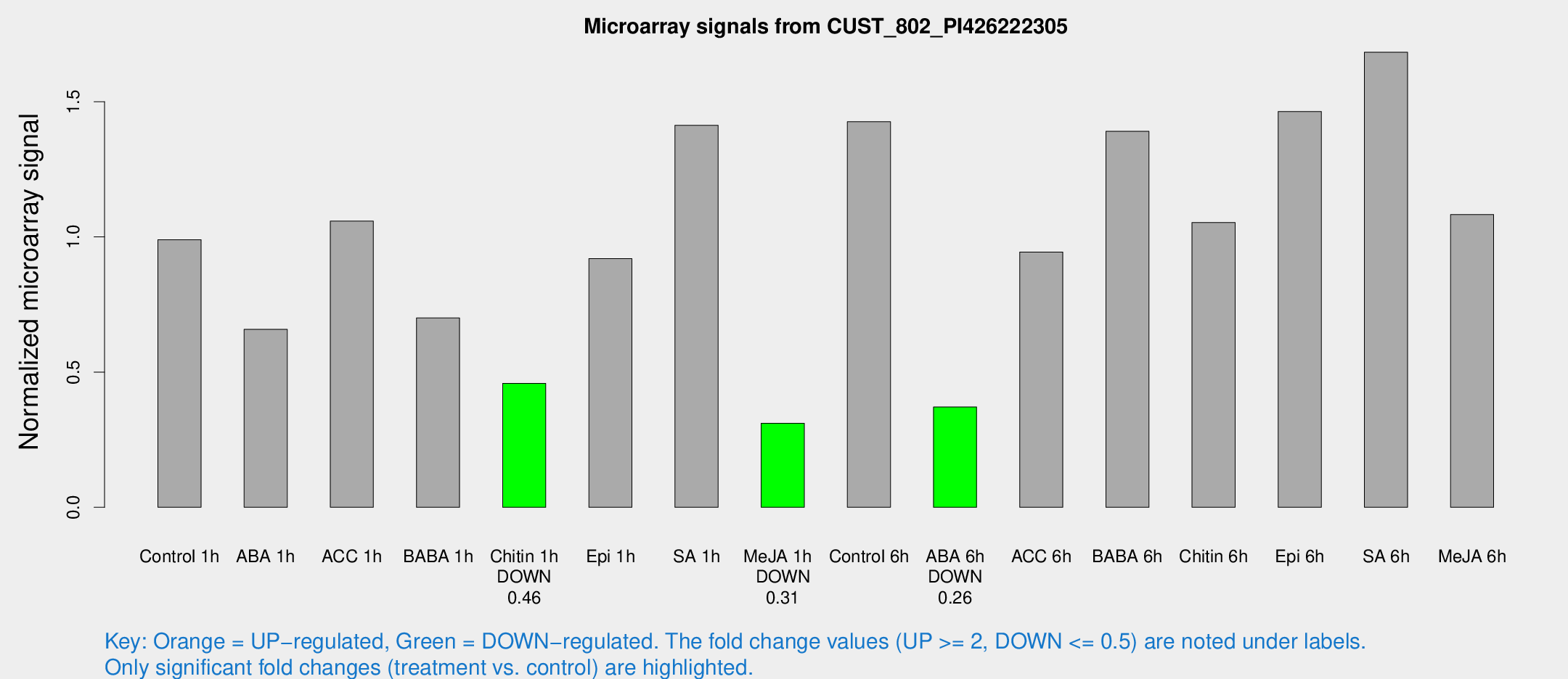
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 255.186 | 65.7645 | 0.989749 | 0.182362 |
| ABA 1h | 145.04 | 22.5539 | 0.657968 | 0.0794864 |
| ACC 1h | 295.147 | 84.1719 | 1.05837 | 0.321193 |
| BABA 1h | 172.657 | 36.8396 | 0.700075 | 0.0986068 |
| Chitin 1h | 102.831 | 17.4946 | 0.458383 | 0.0418187 |
| Epi 1h | 195.2 | 20.6115 | 0.919714 | 0.0818739 |
| SA 1h | 359.062 | 53.8661 | 1.41236 | 0.136167 |
| Me-JA 1h | 62.7732 | 9.42493 | 0.310563 | 0.0313508 |
| Control 6h | 361.44 | 68.9722 | 1.42581 | 0.181971 |
| ABA 6h | 98.8733 | 18.6195 | 0.371036 | 0.0474059 |
| ACC 6h | 263.566 | 16.755 | 0.944092 | 0.0806388 |
| BABA 6h | 378.184 | 22.172 | 1.39034 | 0.0813188 |
| Chitin 6h | 272.494 | 16.1584 | 1.053 | 0.0622663 |
| Epi 6h | 402.964 | 34.1193 | 1.46358 | 0.0855278 |
| SA 6h | 415.684 | 67.6243 | 1.68286 | 0.105277 |
| Me-JA 6h | 284.732 | 87.76 | 1.08264 | 0.242123 |
Source Transcript PGSC0003DMT400003459 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g089080.2 | +2 | 3e-37 | 135 | 73/110 (66%) | genomic_reference:SL2.50ch02 gene_region:45561116-45563543 transcript_region:SL2.50ch02:45561116..45563543+ functional_description:Rapid alkalinization factor 5 (AHRD V1 *-*- Q6DN46_SOLCH) |
| TAIR PP10 | None | - | - | - | - | - |
