Probe CUST_5547_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5547_PI426222305 | JHI_St_60k_v1 | DMT400007053 | ATATGGTGGTGCCGCCGGCGTTGGATGAGAAAGATCCGAATTATGTGAATGAAGAAGAAA |
All Microarray Probes Designed to Gene DMG400002724
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5547_PI426222305 | JHI_St_60k_v1 | DMT400007053 | ATATGGTGGTGCCGCCGGCGTTGGATGAGAAAGATCCGAATTATGTGAATGAAGAAGAAA |
Microarray Signals from CUST_5547_PI426222305
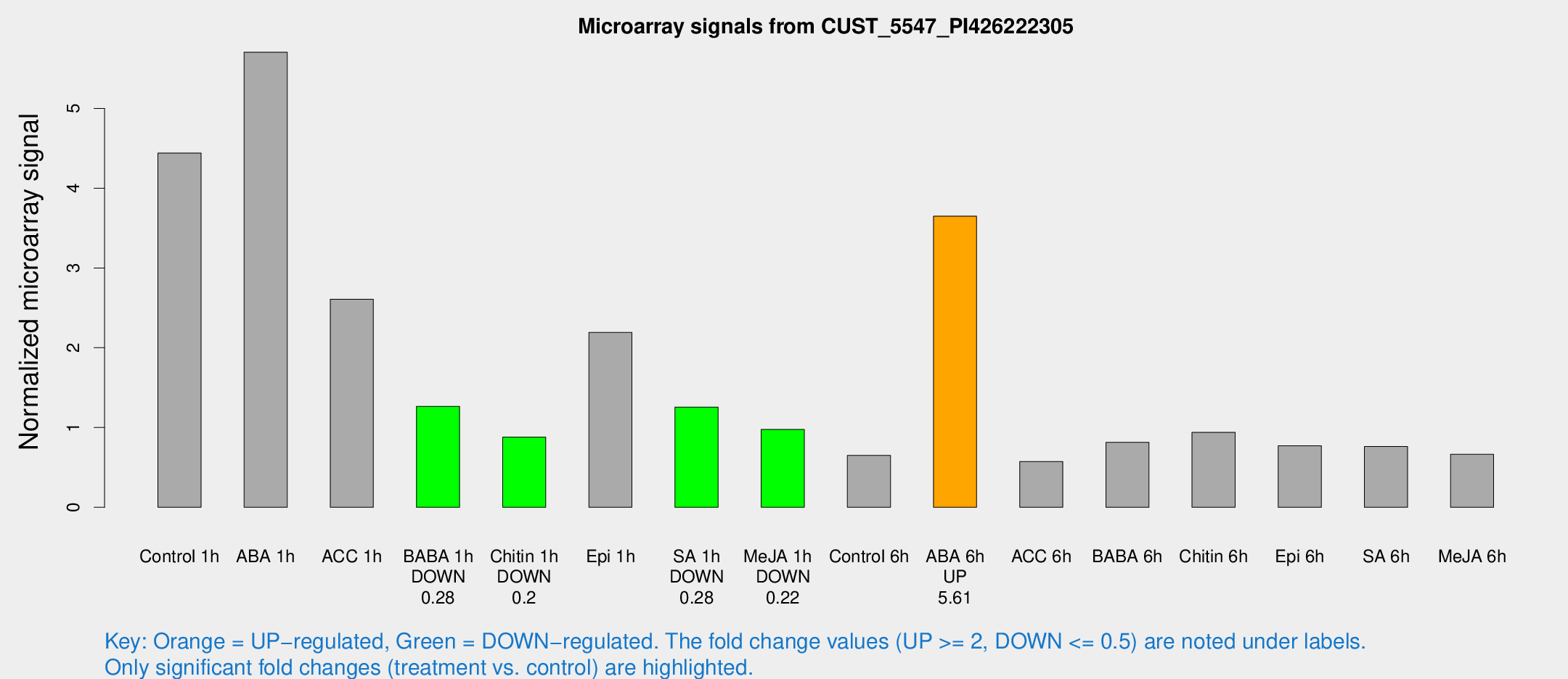
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1522.67 | 198.709 | 4.44027 | 0.538887 |
| ABA 1h | 2148.82 | 1057.75 | 5.70412 | 3.13357 |
| ACC 1h | 1352.57 | 819.818 | 2.60705 | 2.03789 |
| BABA 1h | 417.627 | 49.4095 | 1.26522 | 0.073736 |
| Chitin 1h | 292.093 | 89.9875 | 0.879523 | 0.232935 |
| Epi 1h | 729.597 | 275.03 | 2.19158 | 0.864559 |
| SA 1h | 446.714 | 67.6787 | 1.25639 | 0.139375 |
| Me-JA 1h | 270.259 | 21.0784 | 0.97546 | 0.0671014 |
| Control 6h | 241.8 | 65.2571 | 0.650624 | 0.161096 |
| ABA 6h | 1311.8 | 75.985 | 3.64969 | 0.2109 |
| ACC 6h | 227.656 | 33.9842 | 0.574704 | 0.0453493 |
| BABA 6h | 307.369 | 18.0734 | 0.813979 | 0.0478536 |
| Chitin 6h | 337.33 | 19.7727 | 0.939311 | 0.0550373 |
| Epi 6h | 294.393 | 19.5426 | 0.770881 | 0.0454331 |
| SA 6h | 264.401 | 50.9185 | 0.762361 | 0.194128 |
| Me-JA 6h | 225.936 | 27.1356 | 0.663382 | 0.0393831 |
Source Transcript PGSC0003DMT400007053 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G10020.1 | +2 | 2e-36 | 129 | 71/143 (50%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: response to oxidative stress, anaerobic respiration; LOCATED IN: cellular_component unknown; EXPRESSED IN: 23 plant structures; EXPRESSED DURING: 13 growth stages; Has 47 Blast hits to 47 proteins in 12 species: Archae - 0; Bacteria - 0; Metazoa - 1; Fungi - 0; Plants - 46; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:3091225-3091674 REVERSE LENGTH=149 |
