Probe CUST_51605_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51605_PI426222305 | JHI_St_60k_v1 | DMT400005528 | CAAACTCATGACGTACGCATAATGTTAATGTTCTTATTCGCGTATGATGAATCTCATTCT |
All Microarray Probes Designed to Gene DMG400002161
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51605_PI426222305 | JHI_St_60k_v1 | DMT400005528 | CAAACTCATGACGTACGCATAATGTTAATGTTCTTATTCGCGTATGATGAATCTCATTCT |
| CUST_51581_PI426222305 | JHI_St_60k_v1 | DMT400005530 | GGTTGTCACCTTCATGTCCTAAGTAGCTTCATTCTTAACCCTAACACTTTTAGTATGCCT |
| CUST_51585_PI426222305 | JHI_St_60k_v1 | DMT400005527 | GGTTGTCACCTTCATGTCCTAAGTAGCTTCATTCTTAACCCTAACACTTTTAGTATGCCT |
| CUST_51590_PI426222305 | JHI_St_60k_v1 | DMT400005529 | GGTTGTCACCTTCATGTCCTAAGTAGCTTCATTCTTAACCCTAACACTTTTAGTATGCCT |
Microarray Signals from CUST_51605_PI426222305
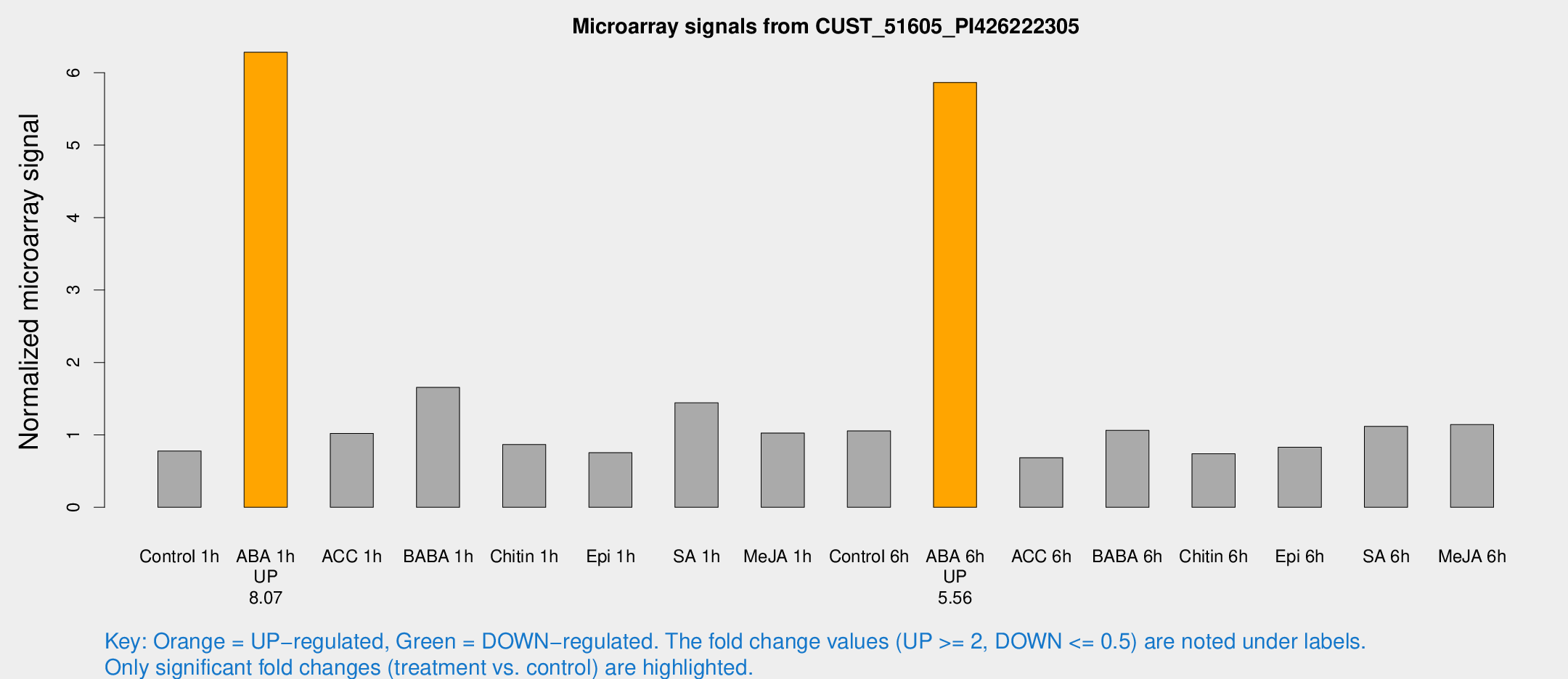
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.09964 | 3.60369 | 0.778134 | 0.403226 |
| ABA 1h | 53.6417 | 12.6735 | 6.28293 | 1.23689 |
| ACC 1h | 9.78779 | 4.68586 | 1.01923 | 0.500852 |
| BABA 1h | 16.5086 | 5.64033 | 1.65527 | 0.538107 |
| Chitin 1h | 7.47762 | 3.59447 | 0.866324 | 0.444079 |
| Epi 1h | 5.94049 | 3.45894 | 0.753992 | 0.437076 |
| SA 1h | 14.5031 | 3.80172 | 1.44197 | 0.443338 |
| Me-JA 1h | 7.7898 | 3.7281 | 1.02612 | 0.514918 |
| Control 6h | 9.85663 | 3.79919 | 1.05482 | 0.44095 |
| ABA 6h | 62.9436 | 17.7965 | 5.86492 | 1.97144 |
| ACC 6h | 7.2902 | 4.33088 | 0.684756 | 0.396672 |
| BABA 6h | 11.3536 | 4.22071 | 1.06252 | 0.437394 |
| Chitin 6h | 7.13533 | 4.14947 | 0.738793 | 0.429088 |
| Epi 6h | 8.66948 | 4.44157 | 0.830238 | 0.428252 |
| SA 6h | 11.5898 | 4.64969 | 1.11787 | 0.520856 |
| Me-JA 6h | 10.4354 | 3.77881 | 1.142 | 0.42654 |
Source Transcript PGSC0003DMT400005528 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G02020.1 | +2 | 2e-22 | 95 | 50/102 (49%) | Encodes a protein involved in salt tolerance, names SIS (Salt Induced Serine rich). | chr5:386557-387921 REVERSE LENGTH=149 |
