Probe CUST_51280_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51280_PI426222305 | JHI_St_60k_v1 | DMT400016882 | TGGATTTTAATTGATCTAATTCATACCCATGGGAACTTTACCGCACATTTATTGGTTCTG |
All Microarray Probes Designed to Gene DMG400006604
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51291_PI426222305 | JHI_St_60k_v1 | DMT400016881 | TGGATTTTAATTGATCTAATTCATACCCATGGGAACTTTACCGCACATTTATTGGTTCTG |
| CUST_51280_PI426222305 | JHI_St_60k_v1 | DMT400016882 | TGGATTTTAATTGATCTAATTCATACCCATGGGAACTTTACCGCACATTTATTGGTTCTG |
| CUST_51285_PI426222305 | JHI_St_60k_v1 | DMT400016883 | TGGATTTTAATTGATCTAATTCATACCCATGGGAACTTTACCGCACATTTATTGGTTCTG |
| CUST_51295_PI426222305 | JHI_St_60k_v1 | DMT400016880 | GTGCTGGGACCATTTGTGTTGTATGAAACATTTTACCTTTTTACTTCTGCTTCTTTTGCA |
Microarray Signals from CUST_51280_PI426222305
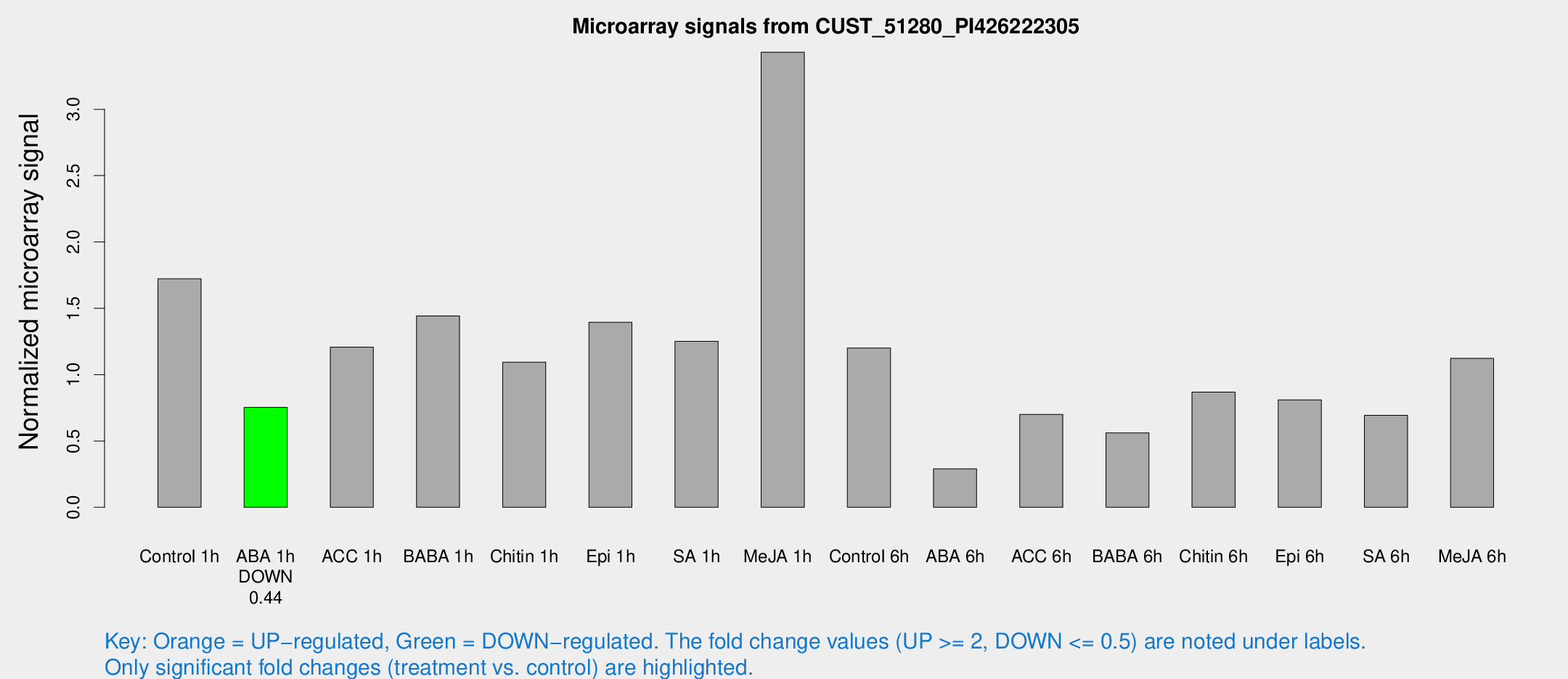
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 509.186 | 108.009 | 1.72227 | 0.334058 |
| ABA 1h | 189.653 | 15.5163 | 0.754107 | 0.0565759 |
| ACC 1h | 379.451 | 105.306 | 1.20784 | 0.284715 |
| BABA 1h | 397.839 | 44.452 | 1.44199 | 0.0841845 |
| Chitin 1h | 278.167 | 16.3936 | 1.09353 | 0.0692978 |
| Epi 1h | 343.1 | 28.106 | 1.39413 | 0.112104 |
| SA 1h | 384.64 | 96.1629 | 1.25156 | 0.33192 |
| Me-JA 1h | 792.566 | 45.9738 | 3.43017 | 0.234662 |
| Control 6h | 361.103 | 84.5802 | 1.20039 | 0.196374 |
| ABA 6h | 90.2527 | 17.0172 | 0.290477 | 0.0376711 |
| ACC 6h | 227.678 | 13.7875 | 0.700484 | 0.0571495 |
| BABA 6h | 184.297 | 36.2365 | 0.56081 | 0.122926 |
| Chitin 6h | 262.773 | 20.8845 | 0.868532 | 0.0516661 |
| Epi 6h | 267.861 | 52.4429 | 0.809332 | 0.199795 |
| SA 6h | 200.492 | 39.0836 | 0.693122 | 0.104074 |
| Me-JA 6h | 323.096 | 50.0438 | 1.12245 | 0.081526 |
Source Transcript PGSC0003DMT400016882 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28550.3 | +1 | 3e-59 | 203 | 123/254 (48%) | related to AP2.7 | chr2:12226168-12228251 REVERSE LENGTH=464 |
