Probe CUST_50720_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50720_PI426222305 | JHI_St_60k_v1 | DMT400071732 | CATTGATTCTGTTCATGGTATTTGCAAAGATTGAGTACATTGAGGAATTTGAATCTCCAC |
All Microarray Probes Designed to Gene DMG400027904
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50729_PI426222305 | JHI_St_60k_v1 | DMT400071728 | CATTGATTCTGTTCATGGTATTTGCAAAGATTGAGTACATTGAGGAATTTGAATCTCCAC |
| CUST_50726_PI426222305 | JHI_St_60k_v1 | DMT400071734 | GATCTTGAGACAATGAATGCTCGTGCAGGGAACAGCATTAGGGAAGGGTATAGAAGTTGA |
| CUST_50720_PI426222305 | JHI_St_60k_v1 | DMT400071732 | CATTGATTCTGTTCATGGTATTTGCAAAGATTGAGTACATTGAGGAATTTGAATCTCCAC |
| CUST_50728_PI426222305 | JHI_St_60k_v1 | DMT400071731 | CATTGATTCTGTTCATGGTATTTGCAAAGATTGAGTACATTGAGGAATTTGAATCTCCAC |
Microarray Signals from CUST_50720_PI426222305
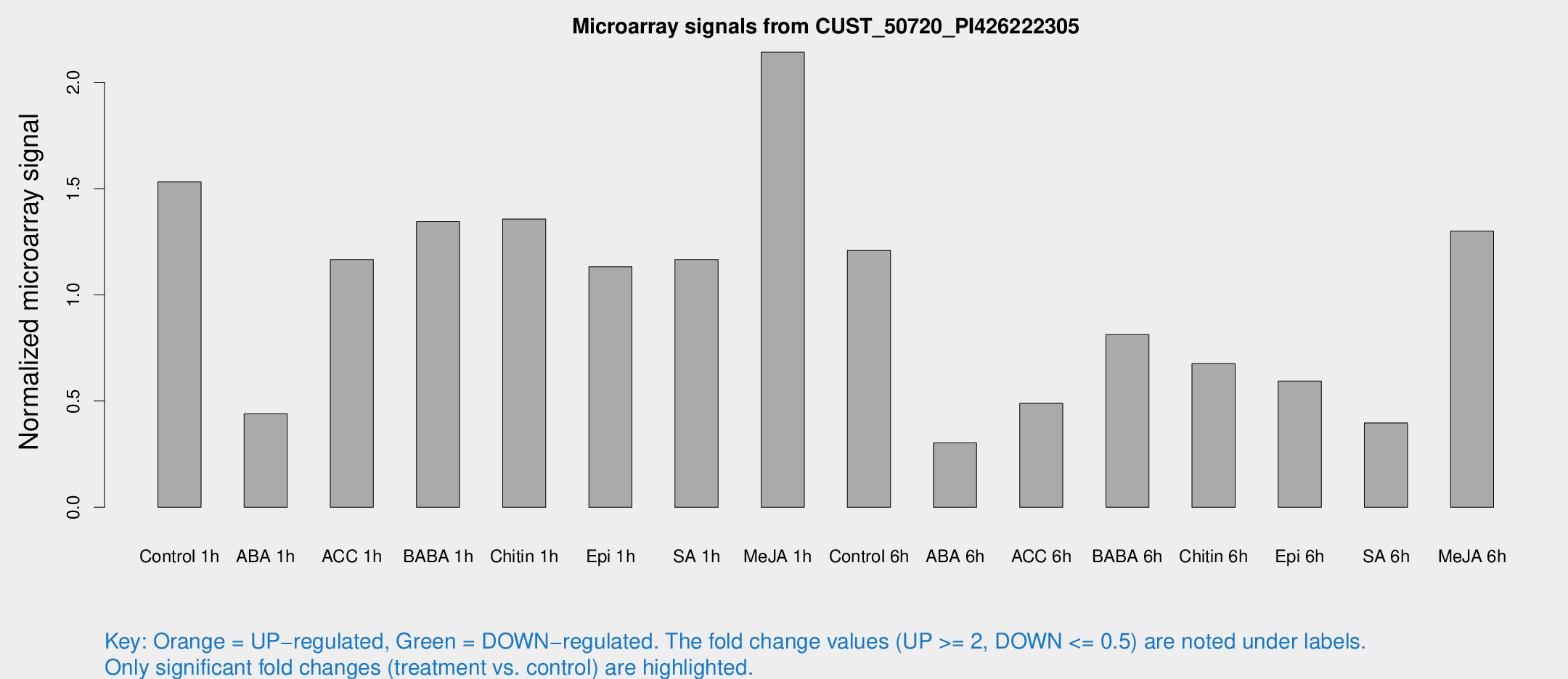
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 67.3514 | 11.9958 | 1.53166 | 0.194125 |
| ABA 1h | 20.669 | 7.9617 | 0.439856 | 0.26721 |
| ACC 1h | 54.3671 | 12.1083 | 1.16606 | 0.219625 |
| BABA 1h | 58.7946 | 13.1188 | 1.3447 | 0.209677 |
| Chitin 1h | 53.7944 | 9.65974 | 1.35654 | 0.324747 |
| Epi 1h | 42.196 | 4.27759 | 1.13189 | 0.115339 |
| SA 1h | 53.6176 | 11.4338 | 1.16588 | 0.316792 |
| Me-JA 1h | 76.5346 | 12.5738 | 2.14168 | 0.178733 |
| Control 6h | 52.6818 | 7.40156 | 1.20879 | 0.136408 |
| ABA 6h | 15.1223 | 4.19089 | 0.303002 | 0.107983 |
| ACC 6h | 26.0623 | 7.7407 | 0.489258 | 0.097017 |
| BABA 6h | 39.7119 | 5.69709 | 0.812761 | 0.100955 |
| Chitin 6h | 32.5524 | 8.01745 | 0.676383 | 0.161613 |
| Epi 6h | 35.9103 | 13.2765 | 0.594582 | 0.470557 |
| SA 6h | 18.5673 | 5.14143 | 0.397124 | 0.179334 |
| Me-JA 6h | 58.9673 | 15.7524 | 1.30042 | 0.246277 |
Source Transcript PGSC0003DMT400071732 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28550.3 | +1 | 6e-106 | 332 | 241/469 (51%) | related to AP2.7 | chr2:12226168-12228251 REVERSE LENGTH=464 |
