Probe CUST_50694_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50694_PI426222305 | JHI_St_60k_v1 | DMT400022145 | TCATCGTCCGATCCATTTGTGAAAGTATATGGTGAACAACCTTATAGTTACTATGGAAAA |
All Microarray Probes Designed to Gene DMG400008591
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50677_PI426222305 | JHI_St_60k_v1 | DMT400022146 | CTGCAAGGGTGTCTACATCGATCATTTCTTCACGATTCTTGAATTTTACAAGTAATGAAT |
| CUST_50694_PI426222305 | JHI_St_60k_v1 | DMT400022145 | TCATCGTCCGATCCATTTGTGAAAGTATATGGTGAACAACCTTATAGTTACTATGGAAAA |
| CUST_50688_PI426222305 | JHI_St_60k_v1 | DMT400022143 | GTGATATGTTTGATTACATACCAAAGGGGGATGCTATTTTCATGAAGTGGGTGCTAACAA |
| CUST_50680_PI426222305 | JHI_St_60k_v1 | DMT400022144 | CGGTGTTGTAGTTGTAAAATAGAACTACGTTTTCTACTTTATATGAGGATGGAAGATTGG |
| CUST_50685_PI426222305 | JHI_St_60k_v1 | DMT400022142 | TGATTACATACCAAAGGGGGATGCTATTTTCATGAAGTGGATACTAGCAATATTTACAGA |
Microarray Signals from CUST_50694_PI426222305
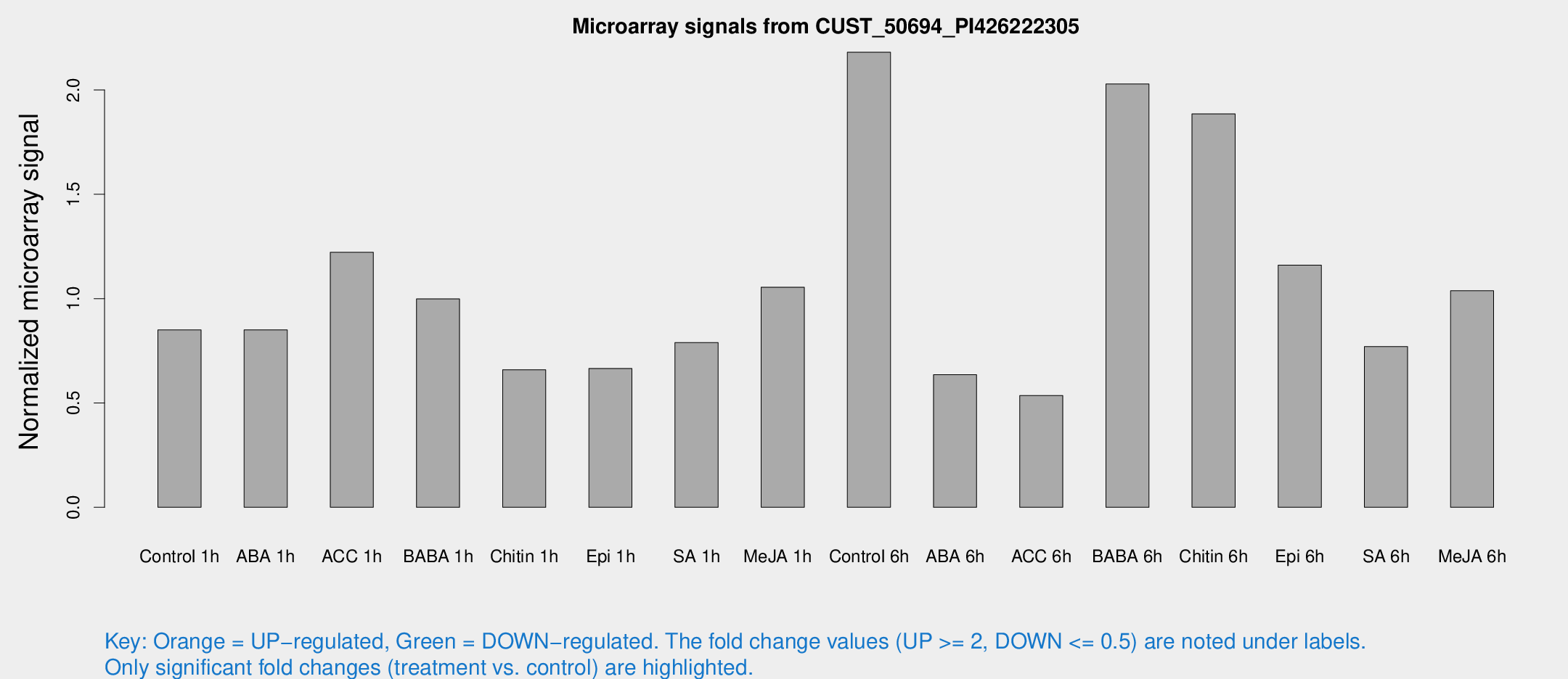
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 43.2034 | 12.843 | 0.850157 | 0.188703 |
| ABA 1h | 39.6715 | 14.2191 | 0.850457 | 0.250253 |
| ACC 1h | 62.4597 | 14.8272 | 1.22182 | 0.21793 |
| BABA 1h | 45.1828 | 4.08623 | 0.998764 | 0.0903035 |
| Chitin 1h | 36.029 | 16.1212 | 0.659104 | 0.402454 |
| Epi 1h | 29.368 | 8.49011 | 0.66531 | 0.18872 |
| SA 1h | 39.0779 | 6.65444 | 0.789621 | 0.134547 |
| Me-JA 1h | 41.8939 | 8.3633 | 1.0548 | 0.298289 |
| Control 6h | 192.063 | 98.001 | 2.18071 | 3.91616 |
| ABA 6h | 34.3879 | 10.2045 | 0.635243 | 0.220395 |
| ACC 6h | 48.7184 | 30.8702 | 0.535637 | 0.496726 |
| BABA 6h | 107.09 | 8.0075 | 2.0284 | 0.134085 |
| Chitin 6h | 100.543 | 25.8763 | 1.88503 | 0.485279 |
| Epi 6h | 86.5122 | 36.1374 | 1.16028 | 1.40771 |
| SA 6h | 48.3953 | 26.5502 | 0.77017 | 0.516273 |
| Me-JA 6h | 61.6578 | 27.9978 | 1.0377 | 0.508717 |
Source Transcript PGSC0003DMT400022145 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G53140.1 | +1 | 3e-156 | 451 | 223/349 (64%) | O-methyltransferase family protein | chr3:19695692-19697355 FORWARD LENGTH=359 |
