Probe CUST_50621_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50621_PI426222305 | JHI_St_60k_v1 | DMT400065535 | GAATAACCTTCATTTCAATTTTTCACACACACAAGGCTAGGTATGTTAGAAGGGGAAAAA |
All Microarray Probes Designed to Gene DMG400025499
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50621_PI426222305 | JHI_St_60k_v1 | DMT400065535 | GAATAACCTTCATTTCAATTTTTCACACACACAAGGCTAGGTATGTTAGAAGGGGAAAAA |
| CUST_50623_PI426222305 | JHI_St_60k_v1 | DMT400065536 | GAATAACCTTCATTTCAATTTTTCACACACACAAGGCTAGGTATGTTAGAAGGGGAAAAA |
| CUST_50614_PI426222305 | JHI_St_60k_v1 | DMT400065538 | GAATAACCTTCATTTCAATTTTTCACACACACAAGGCTAGGTATGTTAGAAGGGGAAAAA |
| CUST_50622_PI426222305 | JHI_St_60k_v1 | DMT400065539 | GAATAACCTTCATTTCAATTTTTCACACACACAAGGCTAGGTATGTTAGAAGGGGAAAAA |
| CUST_50615_PI426222305 | JHI_St_60k_v1 | DMT400065534 | ATATGCACACACAATCCAATCTATTCGAGCTGATCTTCACTATCCATGATCAGGCATTTC |
Microarray Signals from CUST_50621_PI426222305
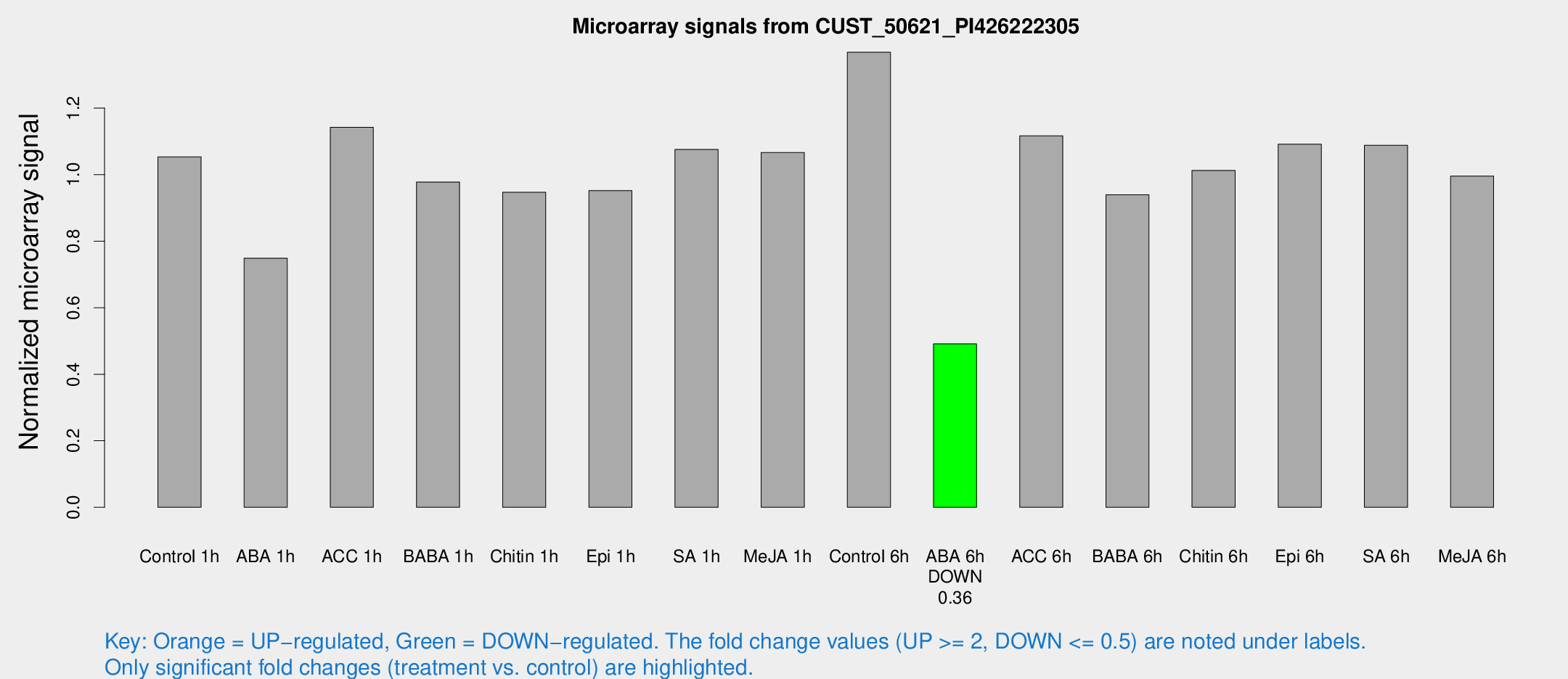
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2591.56 | 348.324 | 1.05364 | 0.0699081 |
| ABA 1h | 1606.21 | 93.0697 | 0.748753 | 0.0432547 |
| ACC 1h | 2930.8 | 511.718 | 1.14202 | 0.137761 |
| BABA 1h | 2344.72 | 371.404 | 0.977855 | 0.0691622 |
| Chitin 1h | 2079.64 | 213.766 | 0.947003 | 0.103839 |
| Epi 1h | 1998.18 | 115.767 | 0.952033 | 0.0549871 |
| SA 1h | 2713.11 | 332.775 | 1.07567 | 0.108978 |
| Me-JA 1h | 2146.9 | 312.128 | 1.06638 | 0.0664106 |
| Control 6h | 3454.86 | 664.416 | 1.36791 | 0.158261 |
| ABA 6h | 1274.63 | 122.773 | 0.491531 | 0.0318145 |
| ACC 6h | 3127.87 | 300.074 | 1.11684 | 0.102851 |
| BABA 6h | 2582.23 | 317.875 | 0.939563 | 0.0805313 |
| Chitin 6h | 2607.64 | 150.701 | 1.01265 | 0.0584829 |
| Epi 6h | 3020.68 | 378.448 | 1.09131 | 0.181045 |
| SA 6h | 2609.73 | 151.113 | 1.08841 | 0.0747484 |
| Me-JA 6h | 2437.34 | 303.237 | 0.995751 | 0.0575073 |
Source Transcript PGSC0003DMT400065535 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05200.1 | +3 | 0.0 | 860 | 424/651 (65%) | glutamate receptor 3.4 | chr1:1505642-1509002 FORWARD LENGTH=959 |
