Probe CUST_4970_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4970_PI426222305 | JHI_St_60k_v1 | DMT400009261 | TGCACTCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCG |
All Microarray Probes Designed to Gene DMG400003593
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4970_PI426222305 | JHI_St_60k_v1 | DMT400009261 | TGCACTCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCG |
| CUST_5318_PI426222305 | JHI_St_60k_v1 | DMT400009260 | TGCACTCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCG |
| CUST_5387_PI426222305 | JHI_St_60k_v1 | DMT400009257 | TCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCGTCTAA |
| CUST_5020_PI426222305 | JHI_St_60k_v1 | DMT400009259 | TCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCGTCTAA |
| CUST_5434_PI426222305 | JHI_St_60k_v1 | DMT400009258 | TCATAGTAATGGTGGTAAAATTACCCCATGAAAACATAATCTTACTTGTGAGCCGTCTAA |
| CUST_5312_PI426222305 | JHI_St_60k_v1 | DMT400009256 | TCGCCGTTTCAAGAATCCAACTATTTTTCTTAAGATTTCCTGTGATGCAGAATATGTCTT |
Microarray Signals from CUST_4970_PI426222305
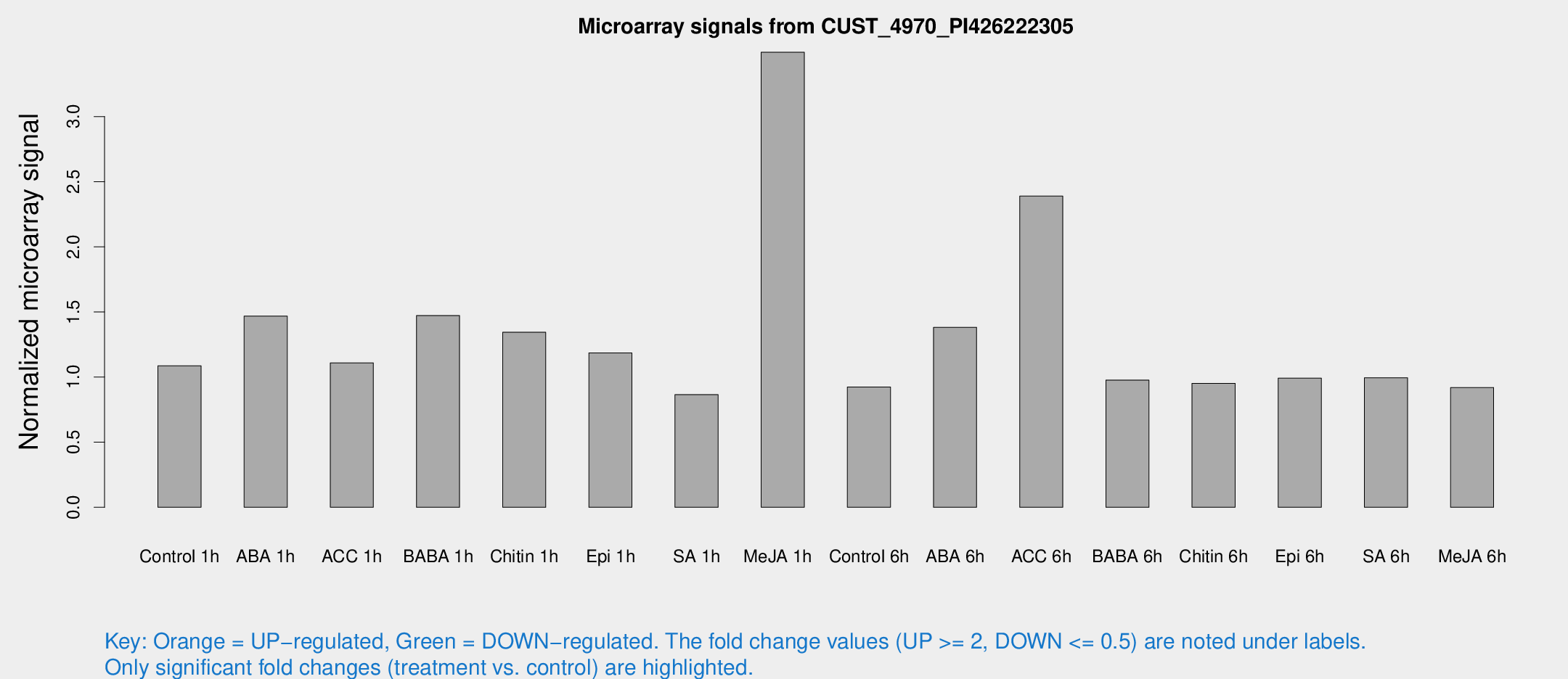
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.15 | 3.64553 | 1.0867 | 0.535538 |
| ABA 1h | 9.70183 | 3.70646 | 1.46832 | 0.61196 |
| ACC 1h | 8.36186 | 4.84365 | 1.10886 | 0.618014 |
| BABA 1h | 10.734 | 3.96149 | 1.47235 | 0.616662 |
| Chitin 1h | 8.92426 | 3.84989 | 1.34421 | 0.608811 |
| Epi 1h | 7.49379 | 3.71118 | 1.18574 | 0.602181 |
| SA 1h | 6.36459 | 3.69234 | 0.865236 | 0.501949 |
| Me-JA 1h | 21.4604 | 4.98922 | 3.49387 | 0.714533 |
| Control 6h | 6.62892 | 3.83958 | 0.923488 | 0.534788 |
| ABA 6h | 12.5689 | 5.54782 | 1.38195 | 0.742172 |
| ACC 6h | 39.6839 | 30.8654 | 2.38965 | 4.07705 |
| BABA 6h | 7.93988 | 4.19301 | 0.976573 | 0.523055 |
| Chitin 6h | 7.26667 | 4.21774 | 0.952067 | 0.551808 |
| Epi 6h | 8.09051 | 4.73548 | 0.991528 | 0.57415 |
| SA 6h | 7.04673 | 4.08501 | 0.994242 | 0.575969 |
| Me-JA 6h | 6.56149 | 3.80535 | 0.919189 | 0.532512 |
Source Transcript PGSC0003DMT400009261 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
