Probe CUST_48616_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48616_PI426222305 | JHI_St_60k_v1 | DMT400077529 | TACTCATGCAAGATGTATGATTCTGGAATGCTTCCGCTGTGTATGCTATTTTCTAACAAT |
All Microarray Probes Designed to Gene DMG400030149
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48616_PI426222305 | JHI_St_60k_v1 | DMT400077529 | TACTCATGCAAGATGTATGATTCTGGAATGCTTCCGCTGTGTATGCTATTTTCTAACAAT |
| CUST_48609_PI426222305 | JHI_St_60k_v1 | DMT400077530 | ATCTGTCTAGACATAATTGCTTTGAGCAACTTGAAACATCTGGATCTCAGGGGAAACAAT |
| CUST_48611_PI426222305 | JHI_St_60k_v1 | DMT400077531 | GCCGGTAGATTTATGAATCTGGCACATGAAATAAGTTAAGTGTTGCTCTAAAGGAAATTA |
| CUST_48621_PI426222305 | JHI_St_60k_v1 | DMT400077528 | TACTCATGCAAGATGTATGATTCTGGAATGCTTCCGCTGTGTATGCTATTTTCTAACAAT |
| CUST_48624_PI426222305 | JHI_St_60k_v1 | DMT400077532 | TACTCATGCAAGATGTATGATTCTGGAATGCTTCCGCTGTGTATGCTATTTTCTAACAAT |
Microarray Signals from CUST_48616_PI426222305
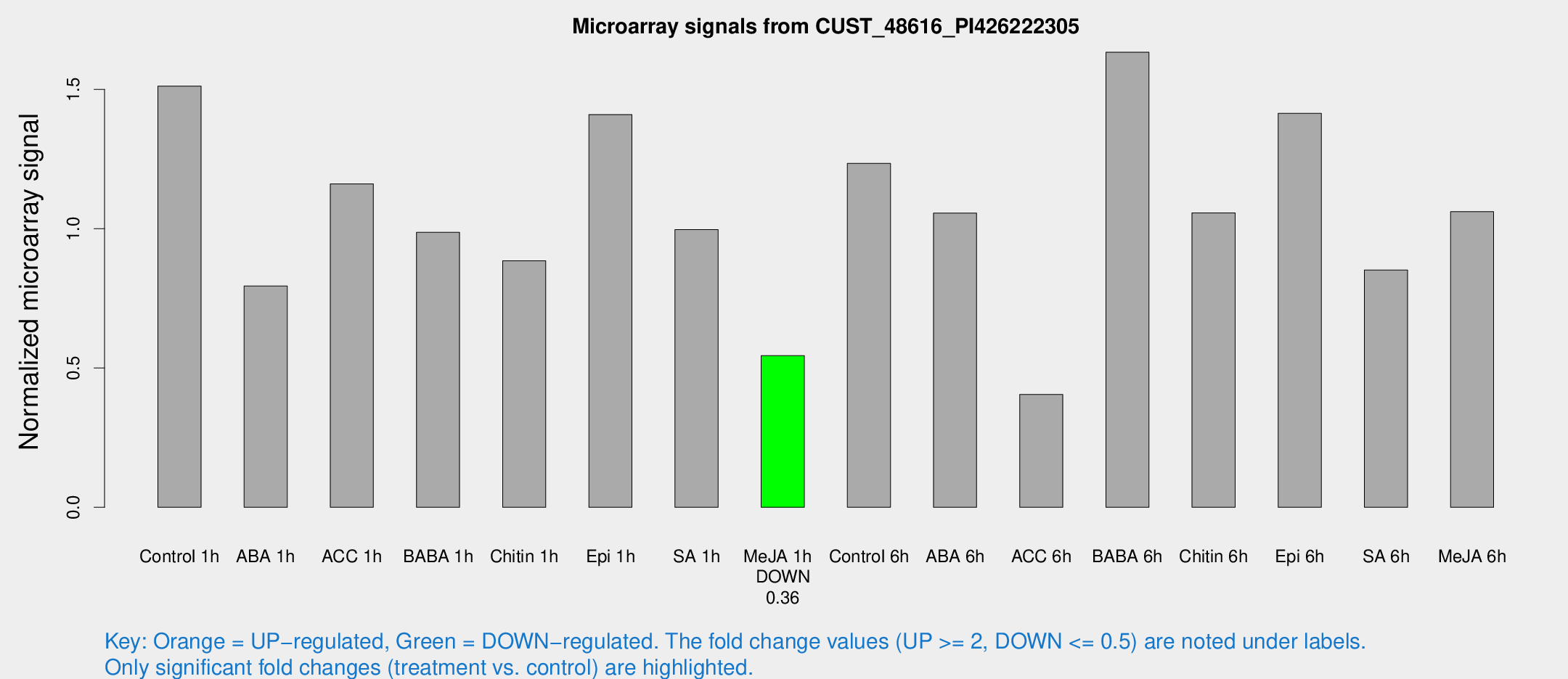
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 24.1173 | 3.98095 | 1.51187 | 0.250311 |
| ABA 1h | 11.2924 | 3.58981 | 0.794112 | 0.258455 |
| ACC 1h | 20.7456 | 5.37349 | 1.16054 | 0.380445 |
| BABA 1h | 17.6644 | 6.64422 | 0.986828 | 0.595046 |
| Chitin 1h | 12.6587 | 3.73426 | 0.884554 | 0.259703 |
| Epi 1h | 20.1192 | 3.77892 | 1.40936 | 0.299496 |
| SA 1h | 17.8124 | 5.12187 | 0.996698 | 0.405724 |
| Me-JA 1h | 7.11583 | 3.83843 | 0.544195 | 0.293714 |
| Control 6h | 20.9732 | 4.69858 | 1.23453 | 0.445268 |
| ABA 6h | 22.4589 | 9.2481 | 1.05579 | 0.615377 |
| ACC 6h | 7.54712 | 4.47618 | 0.40507 | 0.235087 |
| BABA 6h | 32.1751 | 8.73147 | 1.63327 | 0.530284 |
| Chitin 6h | 21.0818 | 6.90108 | 1.05684 | 0.55235 |
| Epi 6h | 26.3499 | 5.35162 | 1.41393 | 0.302945 |
| SA 6h | 20.1666 | 12.9343 | 0.851262 | 0.997825 |
| Me-JA 6h | 19.5711 | 6.587 | 1.06101 | 0.386122 |
Source Transcript PGSC0003DMT400077529 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G58190.2 | +3 | 6e-16 | 81 | 86/302 (28%) | receptor like protein 9 | chr1:21540720-21547996 FORWARD LENGTH=1029 |
