Probe CUST_48448_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48448_PI426222305 | JHI_St_60k_v1 | DMT400055832 | GAACAAAGAGGGATTTTGCTTTCTTATGTGTTGTATGAAATTGATTGGTGGGAGATTAGT |
All Microarray Probes Designed to Gene DMG400021686
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48448_PI426222305 | JHI_St_60k_v1 | DMT400055832 | GAACAAAGAGGGATTTTGCTTTCTTATGTGTTGTATGAAATTGATTGGTGGGAGATTAGT |
Microarray Signals from CUST_48448_PI426222305
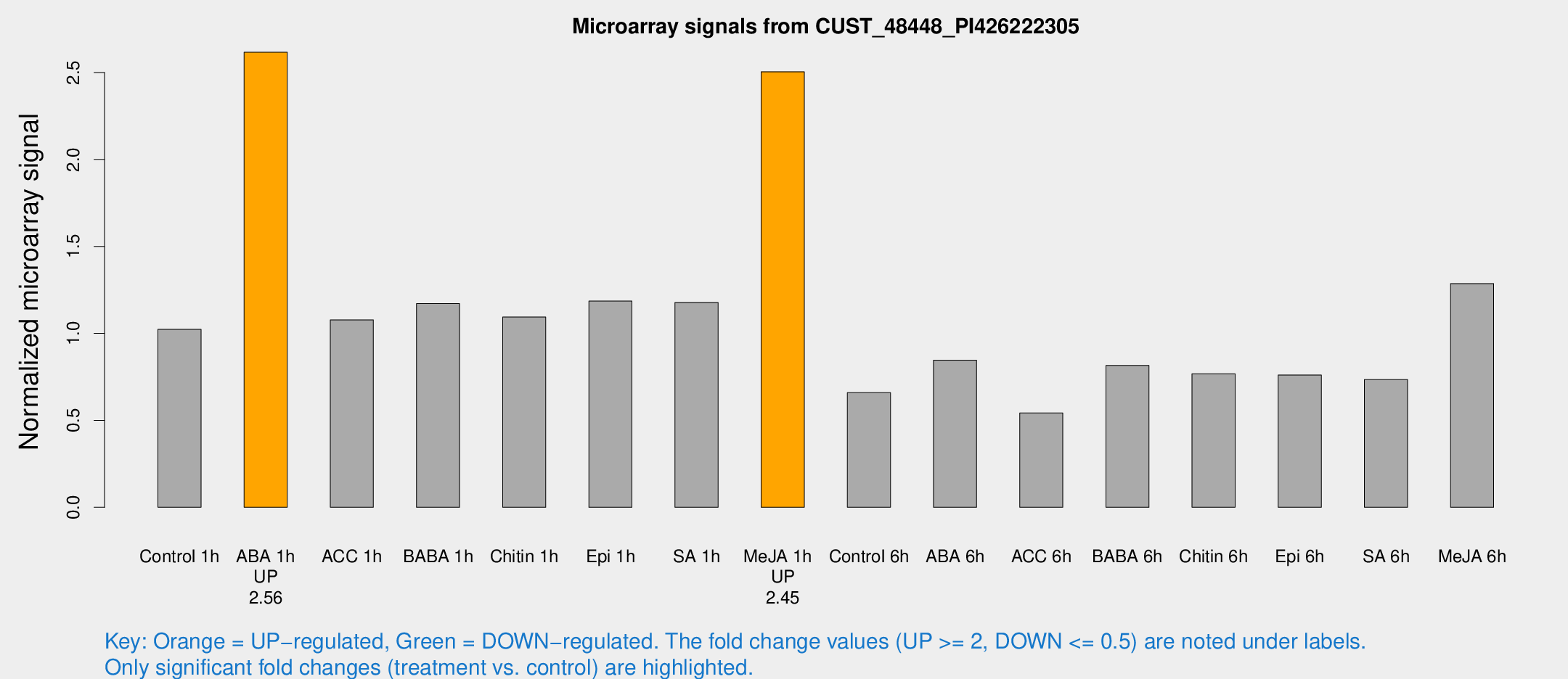
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 604.585 | 58.791 | 1.02317 | 0.0593348 |
| ABA 1h | 1413.93 | 269.751 | 2.61674 | 0.452297 |
| ACC 1h | 648.468 | 37.6498 | 1.07748 | 0.0624714 |
| BABA 1h | 676.403 | 98.0669 | 1.1713 | 0.0679298 |
| Chitin 1h | 587.933 | 88.5707 | 1.09394 | 0.1734 |
| Epi 1h | 603.043 | 39.6435 | 1.1857 | 0.0973819 |
| SA 1h | 724.799 | 104.204 | 1.17785 | 0.130395 |
| Me-JA 1h | 1207.2 | 112.164 | 2.50421 | 0.14475 |
| Control 6h | 441.832 | 150.406 | 0.658937 | 0.201677 |
| ABA 6h | 533.73 | 64.7288 | 0.84568 | 0.0784159 |
| ACC 6h | 370.104 | 45.9091 | 0.542218 | 0.0477297 |
| BABA 6h | 540.693 | 61.2903 | 0.81542 | 0.0634504 |
| Chitin 6h | 478.925 | 27.9358 | 0.768133 | 0.0479286 |
| Epi 6h | 515.19 | 78.68 | 0.760732 | 0.177541 |
| SA 6h | 440.397 | 81.7521 | 0.734331 | 0.0771044 |
| Me-JA 6h | 785.27 | 160.293 | 1.28662 | 0.176665 |
Source Transcript PGSC0003DMT400055832 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G28290.1 | +1 | 1e-06 | 44 | 19/21 (90%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 23 plant structures; EXPRESSED DURING: 15 growth stages; Has 45 Blast hits to 45 proteins in 11 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 45; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr4:14012032-14012464 REVERSE LENGTH=80 |
