Probe CUST_48394_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48394_PI426222305 | JHI_St_60k_v1 | DMT400005781 | ATGAGACTTTTCGACGTGAATCTAAATTTAATCGGACTTCAATATGTGTACCGAACACCC |
All Microarray Probes Designed to Gene DMG400002261
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48394_PI426222305 | JHI_St_60k_v1 | DMT400005781 | ATGAGACTTTTCGACGTGAATCTAAATTTAATCGGACTTCAATATGTGTACCGAACACCC |
| CUST_48398_PI426222305 | JHI_St_60k_v1 | DMT400005782 | TCTAAAGTATAGAAGATGGCTATTTCAACAAAGCTATGCCTCAATCTCTCCCCACAACCT |
Microarray Signals from CUST_48394_PI426222305
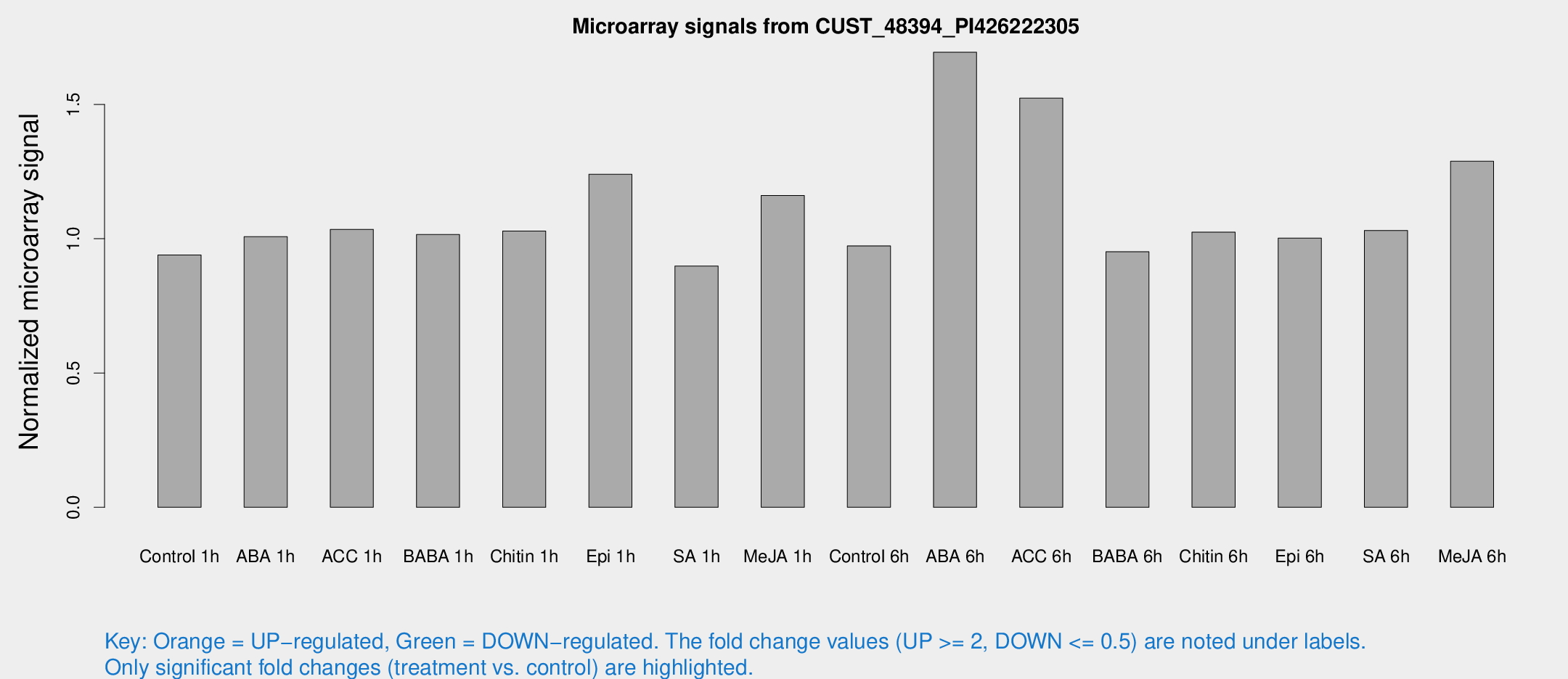
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.56859 | 3.22651 | 0.939581 | 0.544067 |
| ABA 1h | 5.28346 | 3.06273 | 1.00767 | 0.583718 |
| ACC 1h | 6.34459 | 3.70323 | 1.03479 | 0.598958 |
| BABA 1h | 5.80301 | 3.36264 | 1.01573 | 0.588167 |
| Chitin 1h | 5.4933 | 3.19123 | 1.0287 | 0.595487 |
| Epi 1h | 6.45156 | 3.17338 | 1.2399 | 0.63323 |
| SA 1h | 5.46795 | 3.16827 | 0.898145 | 0.520102 |
| Me-JA 1h | 5.63172 | 3.27789 | 1.16095 | 0.673241 |
| Control 6h | 5.7796 | 3.35224 | 0.973321 | 0.564379 |
| ABA 6h | 15.4723 | 9.53506 | 1.69438 | 1.1493 |
| ACC 6h | 11.8926 | 4.50008 | 1.52351 | 0.701943 |
| BABA 6h | 6.32093 | 3.67099 | 0.951292 | 0.551362 |
| Chitin 6h | 6.46185 | 3.74322 | 1.02453 | 0.59329 |
| Epi 6h | 6.76441 | 3.89753 | 1.00248 | 0.574085 |
| SA 6h | 6.04134 | 3.50149 | 1.03086 | 0.597298 |
| Me-JA 6h | 8.41747 | 3.3542 | 1.28842 | 0.61675 |
Source Transcript PGSC0003DMT400005781 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G25625.1 | +2 | 2e-14 | 69 | 47/123 (38%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 10 plant structures; EXPRESSED DURING: LP.06 six leaves visible, LP.04 four leaves visible, 4 anthesis, petal differentiation and expansion stage; Has 24 Blast hits to 24 proteins in 9 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 24; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr2:10906562-10907099 FORWARD LENGTH=152 |
