Probe CUST_47990_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47990_PI426222305 | JHI_St_60k_v1 | DMT400074440 | ACTTGTGAGCTTGGTGGACGCCAAAATCTGCATCGTTTATGGTGATAAACCAAATGCTAC |
All Microarray Probes Designed to Gene DMG400028926
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47990_PI426222305 | JHI_St_60k_v1 | DMT400074440 | ACTTGTGAGCTTGGTGGACGCCAAAATCTGCATCGTTTATGGTGATAAACCAAATGCTAC |
| CUST_47961_PI426222305 | JHI_St_60k_v1 | DMT400074439 | ACTTGTGAGCTTGGTGGACGCCAAAATCTGCATCGTTTATGGTGATAAACCAAATGCTAC |
Microarray Signals from CUST_47990_PI426222305
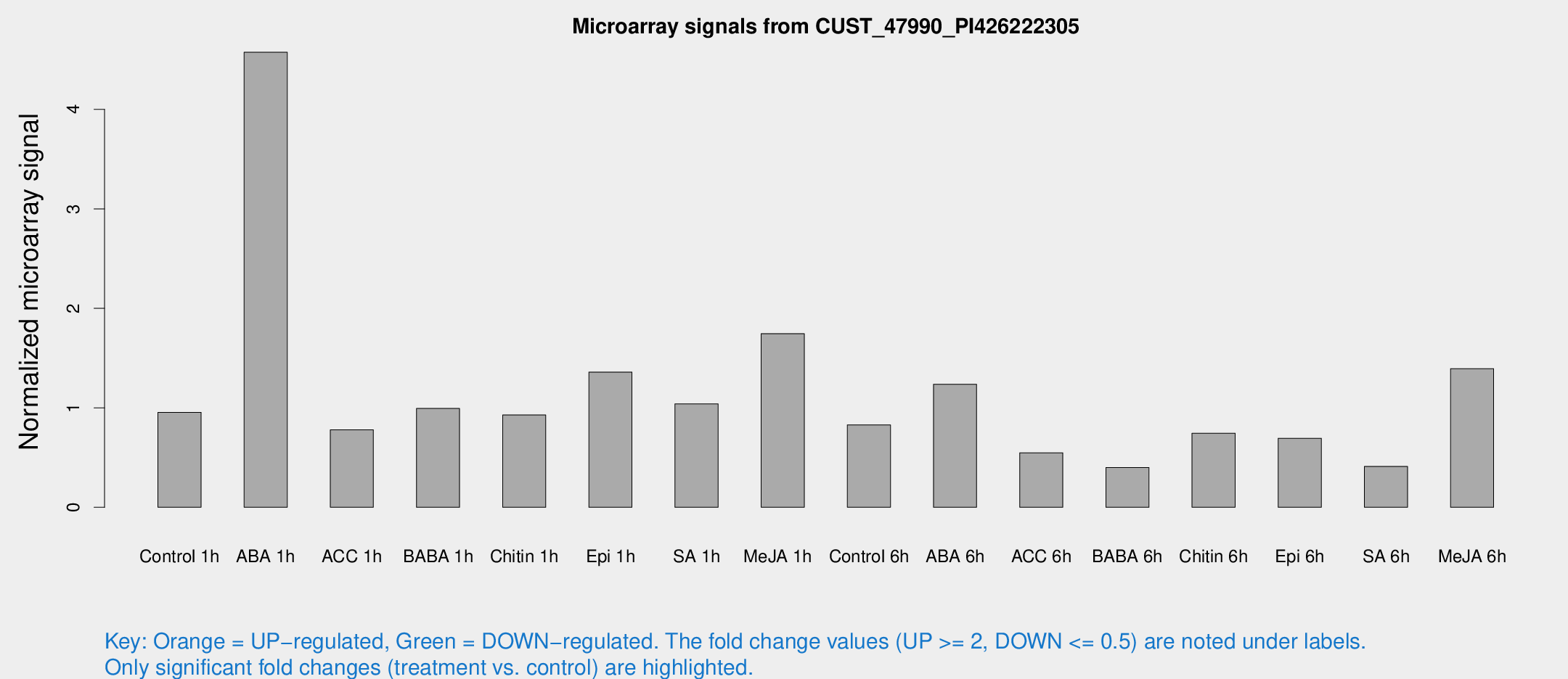
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.8769 | 12.2907 | 0.954843 | 0.256772 |
| ABA 1h | 144.092 | 30.7303 | 4.57463 | 0.793857 |
| ACC 1h | 39.095 | 16.902 | 0.779571 | 0.684728 |
| BABA 1h | 33.2583 | 5.49516 | 0.993823 | 0.186317 |
| Chitin 1h | 29.0591 | 4.87519 | 0.928441 | 0.126651 |
| Epi 1h | 40.1867 | 4.0164 | 1.35951 | 0.137746 |
| SA 1h | 38.1425 | 9.40433 | 1.03951 | 0.251385 |
| Me-JA 1h | 48.182 | 4.45833 | 1.74582 | 0.162971 |
| Control 6h | 29.7433 | 6.45752 | 0.829134 | 0.129502 |
| ABA 6h | 44.8377 | 4.4626 | 1.23723 | 0.125724 |
| ACC 6h | 21.5619 | 4.48139 | 0.548245 | 0.147407 |
| BABA 6h | 17.2175 | 5.65691 | 0.400641 | 0.144519 |
| Chitin 6h | 27.7475 | 4.91422 | 0.74516 | 0.163755 |
| Epi 6h | 26.734 | 4.3208 | 0.694232 | 0.113145 |
| SA 6h | 17.3898 | 8.16262 | 0.411384 | 0.33322 |
| Me-JA 6h | 49.8704 | 11.8377 | 1.3942 | 0.235711 |
Source Transcript PGSC0003DMT400074440 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G43150.1 | +2 | 6e-10 | 57 | 25/61 (41%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: mitochondrion; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 13 growth stages; Has 1807 Blast hits to 1807 proteins in 277 species: Archae - 0; Bacteria - 0; Metazoa - 736; Fungi - 347; Plants - 385; Viruses - 0; Other Eukaryotes - 339 (source: NCBI BLink). | chr5:17325076-17325689 FORWARD LENGTH=91 |
