Probe CUST_47220_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47220_PI426222305 | JHI_St_60k_v1 | DMT400059007 | TCCTAAGGTTAATCTGAGTAATCCCATTGGGAATCAAAGAAAGGAAAAGGGAAATATTGT |
All Microarray Probes Designed to Gene DMG400022916
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47220_PI426222305 | JHI_St_60k_v1 | DMT400059007 | TCCTAAGGTTAATCTGAGTAATCCCATTGGGAATCAAAGAAAGGAAAAGGGAAATATTGT |
Microarray Signals from CUST_47220_PI426222305
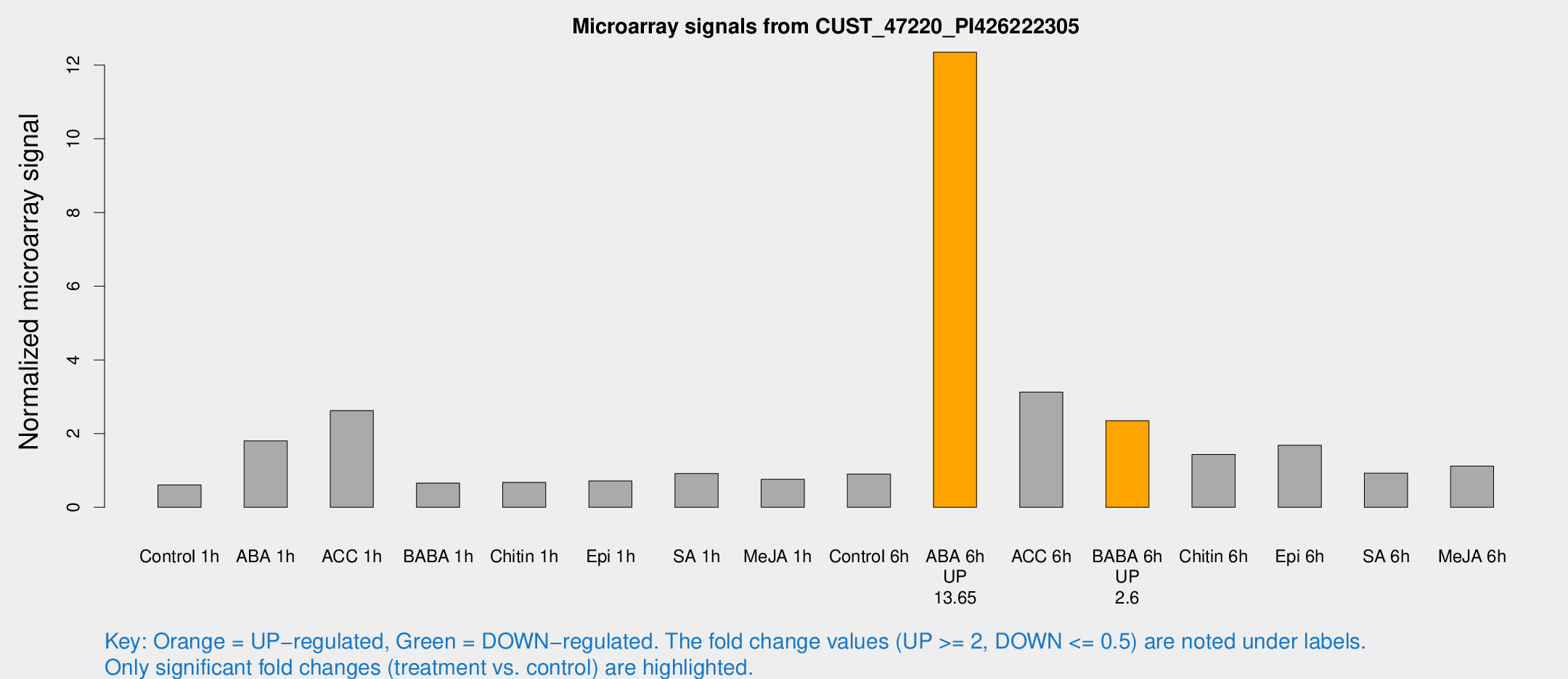
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.42481 | 3.72764 | 0.607433 | 0.351894 |
| ABA 1h | 17.5075 | 3.66404 | 1.802 | 0.404525 |
| ACC 1h | 32.2322 | 11.8824 | 2.62555 | 0.812045 |
| BABA 1h | 6.7005 | 3.8859 | 0.657928 | 0.381426 |
| Chitin 1h | 6.41224 | 3.72167 | 0.673718 | 0.390225 |
| Epi 1h | 6.57103 | 3.63388 | 0.712857 | 0.396328 |
| SA 1h | 12.6422 | 6.39408 | 0.919431 | 0.437986 |
| Me-JA 1h | 6.56628 | 3.81068 | 0.760241 | 0.440466 |
| Control 6h | 10.4399 | 3.85041 | 0.904568 | 0.401925 |
| ABA 6h | 140.9 | 16.9466 | 12.3496 | 1.65669 |
| ACC 6h | 49.6008 | 21.9827 | 3.12896 | 1.65873 |
| BABA 6h | 29.6804 | 7.2887 | 2.34795 | 0.548716 |
| Chitin 6h | 18.4529 | 5.75379 | 1.43418 | 0.696144 |
| Epi 6h | 22.6314 | 6.78159 | 1.6824 | 0.84013 |
| SA 6h | 9.97848 | 4.07872 | 0.926444 | 0.409085 |
| Me-JA 6h | 13.1689 | 4.06916 | 1.11998 | 0.419811 |
Source Transcript PGSC0003DMT400059007 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23690.1 | +1 | 1e-32 | 120 | 88/164 (54%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: N-terminal protein myristoylation; LOCATED IN: cellular_component unknown; EXPRESSED IN: 7 plant structures; EXPRESSED DURING: petal differentiation and expansion stage; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT4G37240.1); Has 243 Blast hits to 243 proteins in 15 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 241; Viruses - 0; Other Eukaryotes - 2 (source: NCBI BLink). | chr2:10072297-10072788 FORWARD LENGTH=163 |
