Probe CUST_47041_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47041_PI426222305 | JHI_St_60k_v1 | DMT400079559 | GTAGCCCCTATGTTCACTTTTATTCATCCACAAAGGATTTTGCACTCTTTGTAAGAAATA |
All Microarray Probes Designed to Gene DMG400030979
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47027_PI426222305 | JHI_St_60k_v1 | DMT400079558 | GTAGCCCCTATGTTCACTTTTATTCATCCACAAAGGATTTTGCACTCTTTGTAAGAAATA |
| CUST_47048_PI426222305 | JHI_St_60k_v1 | DMT400079560 | GTAGCCCCTATGTTCACTTTTATTCATCCACAAAGGATTTTGCACTCTTTGTAAGAAATA |
| CUST_47041_PI426222305 | JHI_St_60k_v1 | DMT400079559 | GTAGCCCCTATGTTCACTTTTATTCATCCACAAAGGATTTTGCACTCTTTGTAAGAAATA |
Microarray Signals from CUST_47041_PI426222305
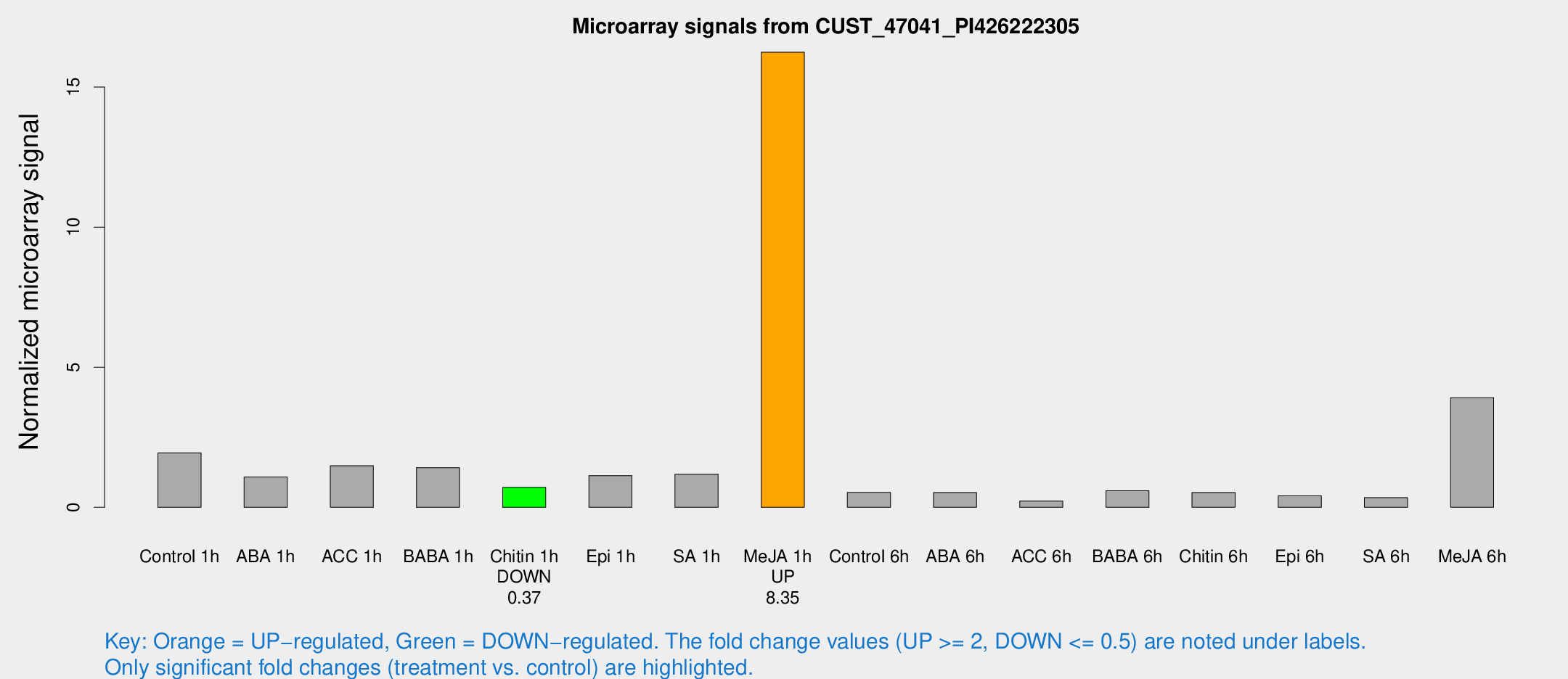
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 103.27 | 33.3613 | 1.9454 | 0.521302 |
| ABA 1h | 51.7043 | 17.573 | 1.08261 | 0.323754 |
| ACC 1h | 85.0612 | 29.6827 | 1.48269 | 0.564417 |
| BABA 1h | 66.063 | 7.21473 | 1.41442 | 0.299377 |
| Chitin 1h | 32.5325 | 7.78055 | 0.712214 | 0.159852 |
| Epi 1h | 47.2757 | 4.53978 | 1.13125 | 0.0997906 |
| SA 1h | 71.0128 | 29.7333 | 1.18109 | 0.722148 |
| Me-JA 1h | 641.004 | 65.5642 | 16.2435 | 2.58274 |
| Control 6h | 40.3011 | 21.688 | 0.532804 | 0.516012 |
| ABA 6h | 35.0904 | 18.7401 | 0.527135 | 0.364583 |
| ACC 6h | 13.8004 | 4.66774 | 0.224573 | 0.0757201 |
| BABA 6h | 32.1373 | 4.12039 | 0.591598 | 0.0777762 |
| Chitin 6h | 28.2402 | 6.51567 | 0.528155 | 0.117632 |
| Epi 6h | 28.0359 | 11.5361 | 0.40791 | 0.338188 |
| SA 6h | 20.6097 | 8.11939 | 0.346102 | 0.26086 |
| Me-JA 6h | 186.918 | 11.3187 | 3.91099 | 0.497846 |
Source Transcript PGSC0003DMT400079559 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G70370.1 | +2 | 0.0 | 696 | 339/599 (57%) | polygalacturonase 2 | chr1:26513003-26514998 REVERSE LENGTH=626 |
