Probe CUST_46113_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46113_PI426222305 | JHI_St_60k_v1 | DMT400014256 | ATGGCGATTTCCTTTGGAAGCATCCATAAGATATTTGGGTAAATGGCATCGTTATAAAAT |
All Microarray Probes Designed to Gene DMG400005592
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46113_PI426222305 | JHI_St_60k_v1 | DMT400014256 | ATGGCGATTTCCTTTGGAAGCATCCATAAGATATTTGGGTAAATGGCATCGTTATAAAAT |
Microarray Signals from CUST_46113_PI426222305
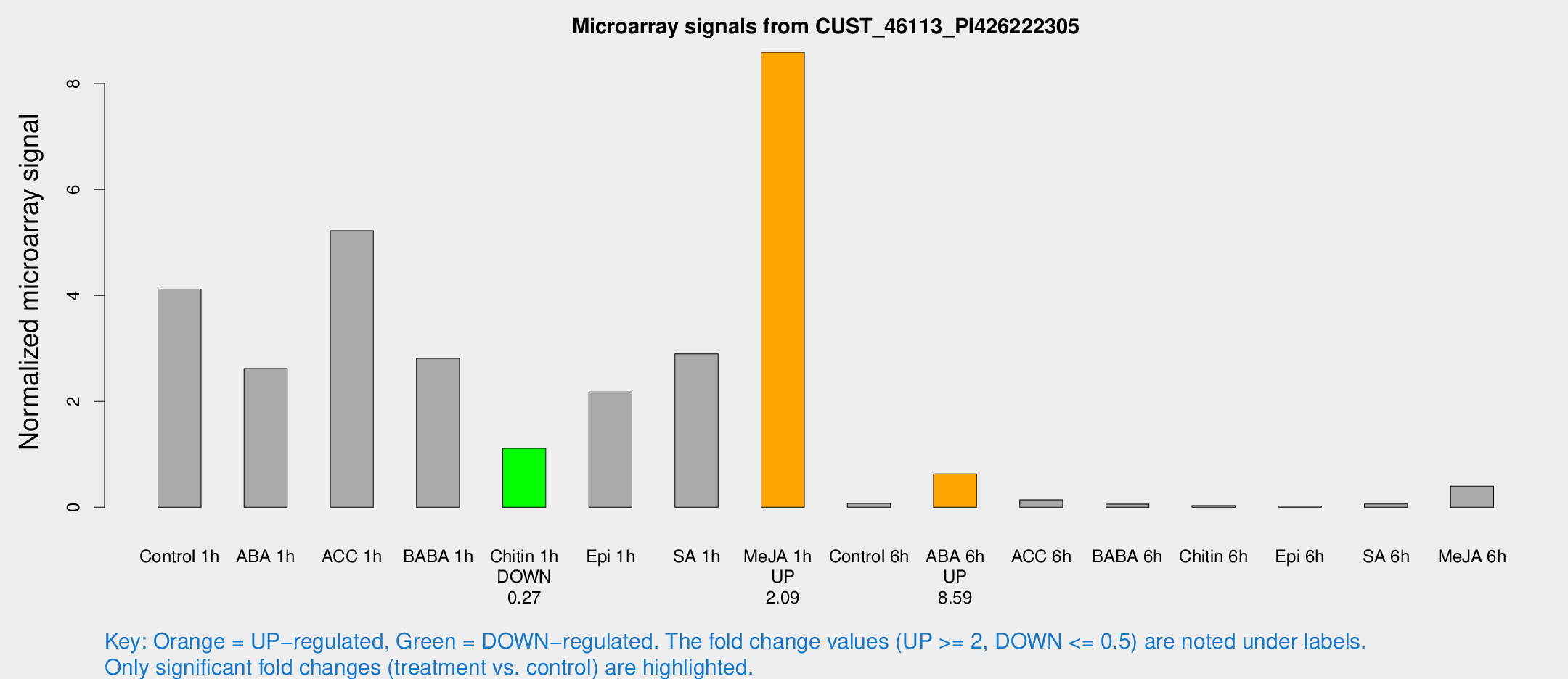
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2561.31 | 602.2 | 4.11832 | 0.665244 |
| ABA 1h | 1582.87 | 502.657 | 2.61867 | 1.05517 |
| ACC 1h | 3869.84 | 1619.74 | 5.22 | 2.35948 |
| BABA 1h | 1658.89 | 324.426 | 2.81106 | 0.4256 |
| Chitin 1h | 607.174 | 90.9206 | 1.11439 | 0.100048 |
| Epi 1h | 1131.63 | 140.392 | 2.17662 | 0.270051 |
| SA 1h | 2081.37 | 848.394 | 2.8965 | 1.4739 |
| Me-JA 1h | 4253.37 | 711.79 | 8.58973 | 0.773408 |
| Control 6h | 56.3066 | 28.162 | 0.0734612 | 0.0360858 |
| ABA 6h | 416.571 | 91.8237 | 0.631273 | 0.126424 |
| ACC 6h | 127.012 | 67.5985 | 0.142674 | 0.109801 |
| BABA 6h | 51.8555 | 26.9583 | 0.0604204 | 0.0342468 |
| Chitin 6h | 23.2603 | 6.42982 | 0.0333519 | 0.0137491 |
| Epi 6h | 20.2196 | 7.18831 | 0.0262454 | 0.0146944 |
| SA 6h | 36.3898 | 3.82587 | 0.0621576 | 0.00973237 |
| Me-JA 6h | 282.49 | 127.149 | 0.399 | 0.238474 |
Source Transcript PGSC0003DMT400014256 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G63380.1 | +3 | 7e-61 | 196 | 103/167 (62%) | NAD(P)-binding Rossmann-fold superfamily protein | chr1:23505582-23506504 FORWARD LENGTH=282 |
