Probe CUST_4397_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4397_PI426222305 | JHI_St_60k_v1 | DMT400047759 | CTTGTTTGGTCTTTTAAAAATACCTTACTTTTGAAGTATCTAAGACACCCCCGATAACAC |
All Microarray Probes Designed to Gene DMG400018565
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4397_PI426222305 | JHI_St_60k_v1 | DMT400047759 | CTTGTTTGGTCTTTTAAAAATACCTTACTTTTGAAGTATCTAAGACACCCCCGATAACAC |
| CUST_4388_PI426222305 | JHI_St_60k_v1 | DMT400047760 | ATTTGGTAGTCGTCTCGTTTCCCTTTCCTGGTGTTCCACATTCACTGAAATCAAGTGTGC |
| CUST_4295_PI426222305 | JHI_St_60k_v1 | DMT400047762 | CCTTGGTCTTGGAGGTGTTGGACTTGGTGTAAGTACATTCGATAGAACTAACGTTTAATG |
Microarray Signals from CUST_4397_PI426222305
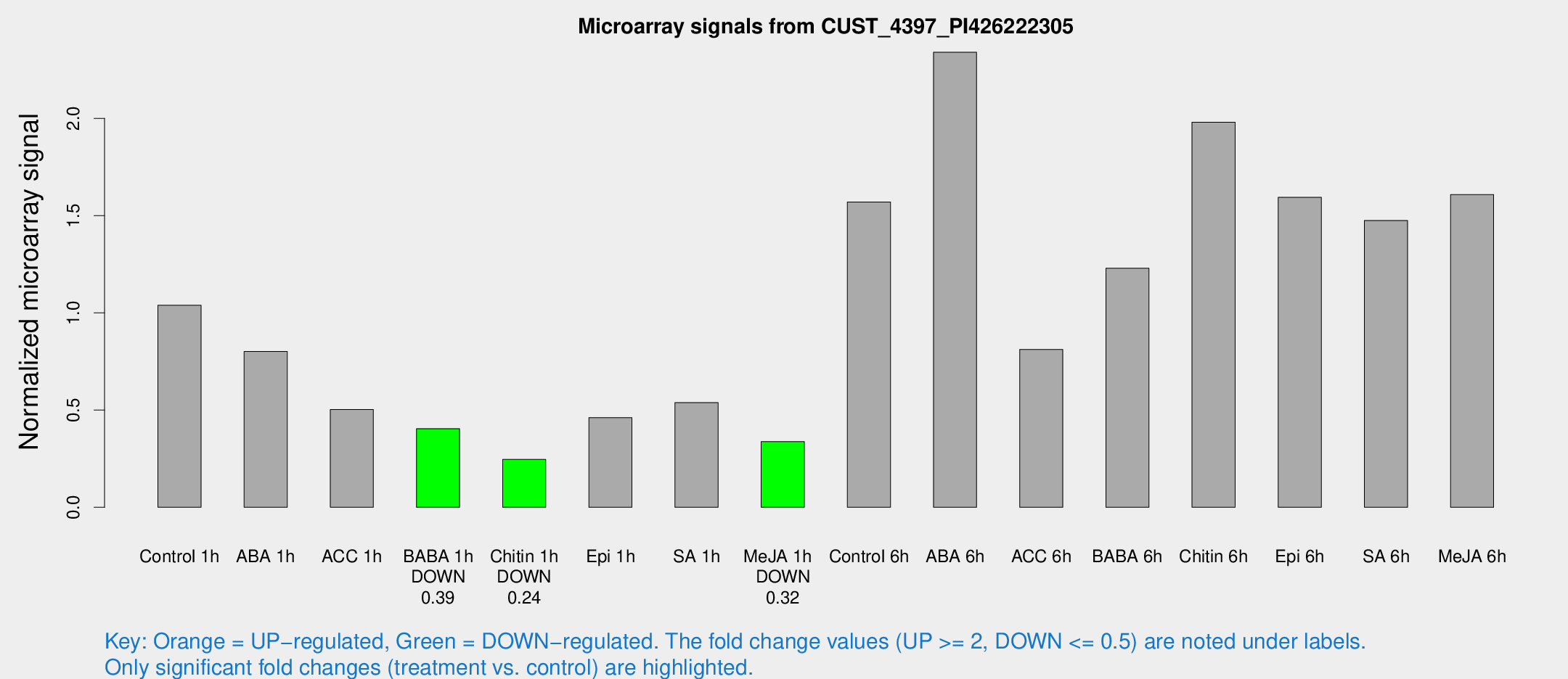
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 24.8873 | 3.24053 | 1.03872 | 0.13898 |
| ABA 1h | 22.2883 | 9.28576 | 0.801365 | 0.792721 |
| ACC 1h | 13.6252 | 3.95886 | 0.502732 | 0.171359 |
| BABA 1h | 9.41717 | 3.11043 | 0.404302 | 0.140603 |
| Chitin 1h | 5.03333 | 2.93222 | 0.246372 | 0.137444 |
| Epi 1h | 10.3057 | 2.95384 | 0.461219 | 0.174653 |
| SA 1h | 16.962 | 8.23451 | 0.538393 | 0.389671 |
| Me-JA 1h | 6.70887 | 3.01382 | 0.337521 | 0.159338 |
| Control 6h | 39.6987 | 10.5889 | 1.56957 | 0.294917 |
| ABA 6h | 61.5921 | 12.5426 | 2.34006 | 0.462335 |
| ACC 6h | 22.0696 | 3.90185 | 0.811738 | 0.143678 |
| BABA 6h | 34.6776 | 9.51466 | 1.22944 | 0.314477 |
| Chitin 6h | 51.3215 | 9.1408 | 1.9801 | 0.451445 |
| Epi 6h | 43.3813 | 6.53647 | 1.59363 | 0.374613 |
| SA 6h | 37.6792 | 11.8789 | 1.4747 | 0.704555 |
| Me-JA 6h | 39.2629 | 8.03861 | 1.60798 | 0.194676 |
Source Transcript PGSC0003DMT400047759 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G32780.1 | +2 | 8e-83 | 265 | 154/355 (43%) | GroES-like zinc-binding dehydrogenase family protein | chr1:11869977-11872595 REVERSE LENGTH=394 |
