Probe CUST_43916_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43916_PI426222305 | JHI_St_60k_v1 | DMT400045385 | CTGCAAACATCAGGTGAGCTTTTTCCTCTTACAATTGAAATCCCAGGGAAGTGTCTTAAC |
All Microarray Probes Designed to Gene DMG400017609
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43916_PI426222305 | JHI_St_60k_v1 | DMT400045385 | CTGCAAACATCAGGTGAGCTTTTTCCTCTTACAATTGAAATCCCAGGGAAGTGTCTTAAC |
| CUST_43922_PI426222305 | JHI_St_60k_v1 | DMT400045386 | AATGATCCTAGACTTTTTCGGATACCATGTTTCCACAGAGGCGTCTCTTGGTGTTGTAGC |
| CUST_43932_PI426222305 | JHI_St_60k_v1 | DMT400045382 | ATTCAAAATGATCCTAGACTTTTTCGAGGCGTCTCTTGGTGTTGTAGCGACATGTCTAAG |
| CUST_43930_PI426222305 | JHI_St_60k_v1 | DMT400045383 | AATGATCCTAGACTTTTTCGGATACCATGTTTCCACAGAGGCGTCTCTTGGTGTTGTAGC |
Microarray Signals from CUST_43916_PI426222305
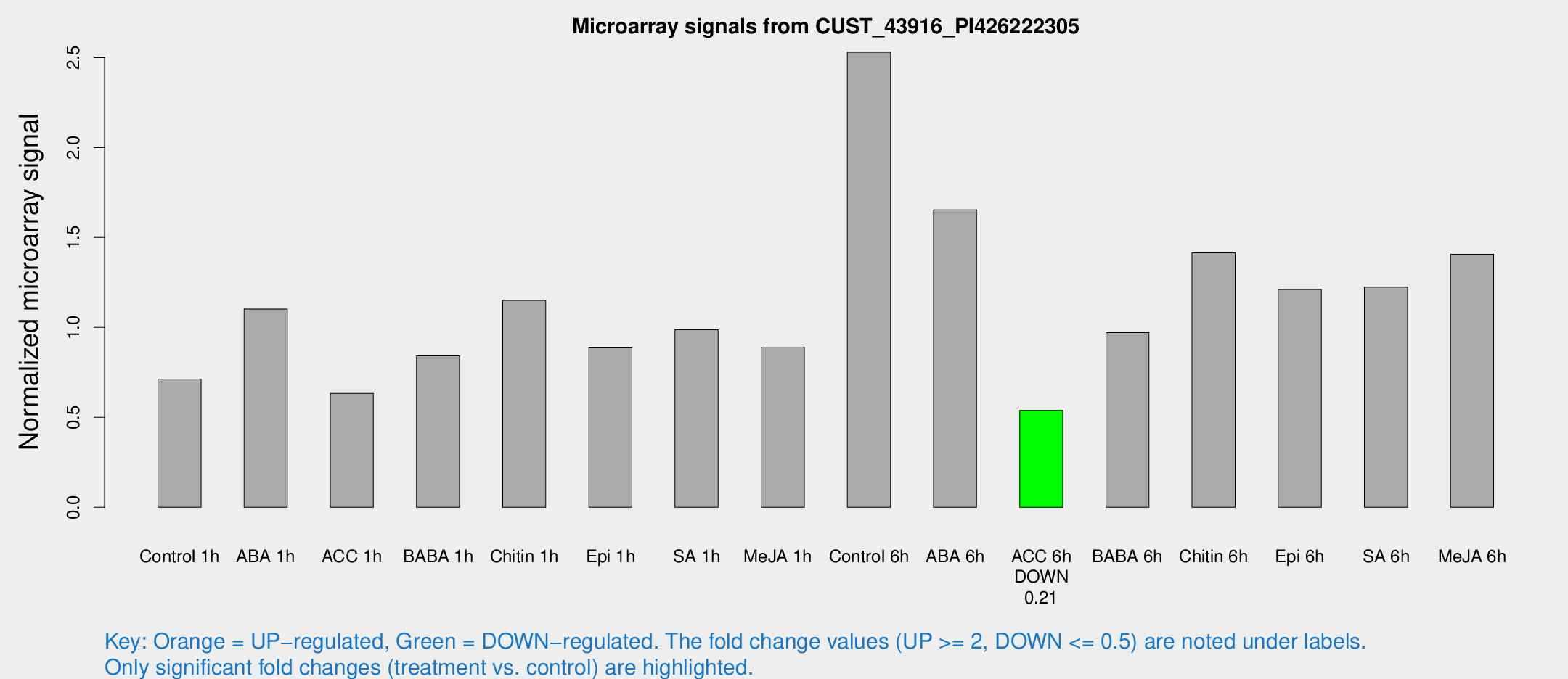
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.81934 | 3.24444 | 0.71338 | 0.291187 |
| ABA 1h | 13.3854 | 4.75541 | 1.10275 | 0.491695 |
| ACC 1h | 8.09321 | 3.51246 | 0.633938 | 0.30942 |
| BABA 1h | 10.2147 | 3.42066 | 0.842053 | 0.329827 |
| Chitin 1h | 13.7526 | 4.8755 | 1.15043 | 0.411286 |
| Epi 1h | 9.71932 | 3.20683 | 0.886436 | 0.341333 |
| SA 1h | 12.5859 | 3.23644 | 0.987597 | 0.28849 |
| Me-JA 1h | 9.08676 | 3.33118 | 0.89034 | 0.364014 |
| Control 6h | 30.5703 | 5.08607 | 2.52981 | 0.345298 |
| ABA 6h | 23.5877 | 8.5673 | 1.65337 | 0.63512 |
| ACC 6h | 7.38071 | 4.08371 | 0.539187 | 0.2923 |
| BABA 6h | 14.984 | 5.63688 | 0.971053 | 0.421421 |
| Chitin 6h | 19.5879 | 5.37432 | 1.4146 | 0.529949 |
| Epi 6h | 17.8426 | 5.09072 | 1.21145 | 0.557353 |
| SA 6h | 19.5531 | 10.9519 | 1.22384 | 0.841558 |
| Me-JA 6h | 21.322 | 10.504 | 1.40682 | 0.705172 |
Source Transcript PGSC0003DMT400045385 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G12130.1 | +1 | 6e-98 | 298 | 151/185 (82%) | integral membrane TerC family protein | chr5:3919613-3922154 FORWARD LENGTH=384 |
