Probe CUST_42108_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42108_PI426222305 | JHI_St_60k_v1 | DMT400067771 | CACATTTGCTAACTTGGCTGGAAAAATTTGCTTATTGGAGATGAAGGTCATGATCAATAT |
All Microarray Probes Designed to Gene DMG400026359
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42108_PI426222305 | JHI_St_60k_v1 | DMT400067771 | CACATTTGCTAACTTGGCTGGAAAAATTTGCTTATTGGAGATGAAGGTCATGATCAATAT |
| CUST_42144_PI426222305 | JHI_St_60k_v1 | DMT400067773 | CACATTTGCTAACTTGGCTGGAAAAATTTGCTTATTGGAGATGAAGGTCATGATCAATAT |
| CUST_42158_PI426222305 | JHI_St_60k_v1 | DMT400067770 | GGAATCTGTCATTTCTTGCTTCGCATGTCGACTATTACCTTATTTTCATCAGTTTCTATG |
Microarray Signals from CUST_42108_PI426222305
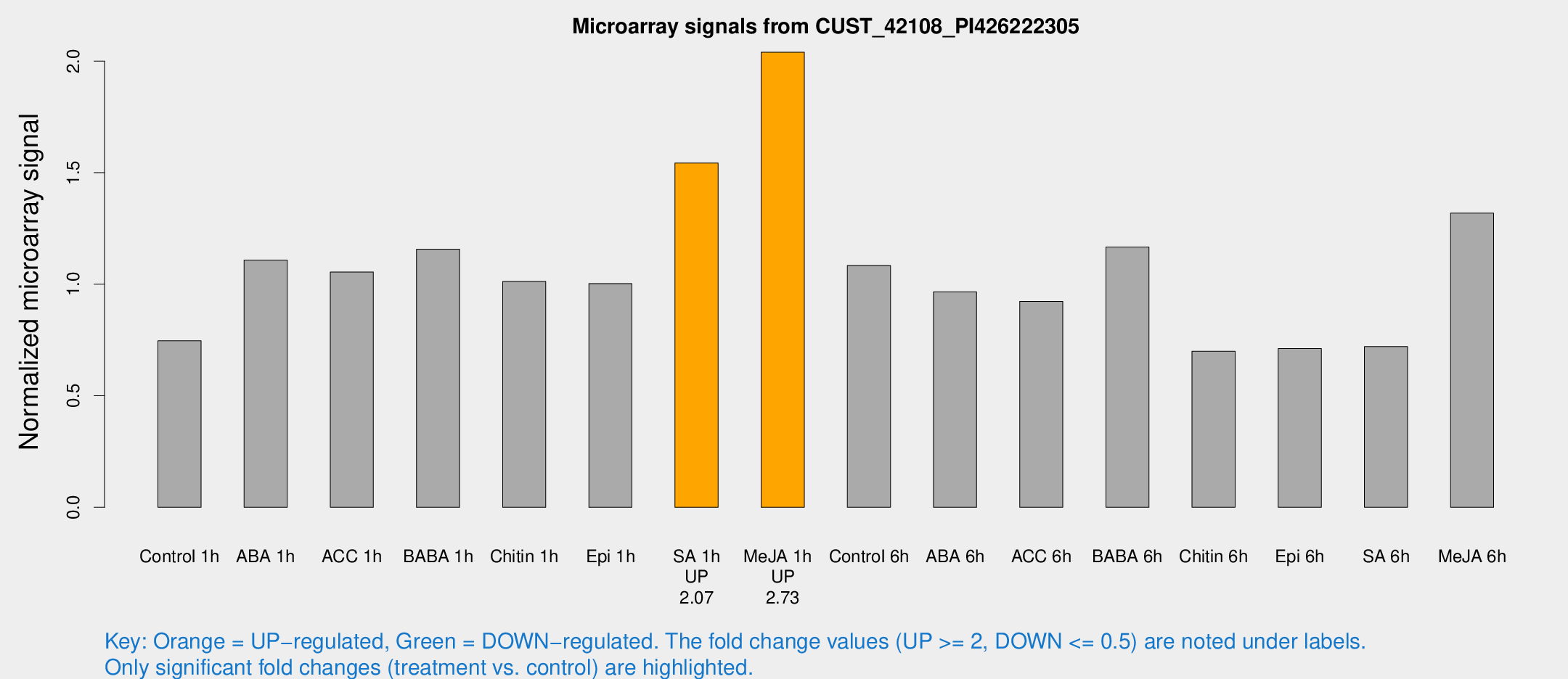
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 946.073 | 74.367 | 0.746737 | 0.0431797 |
| ABA 1h | 1241 | 87.7853 | 1.10825 | 0.0788349 |
| ACC 1h | 1405.85 | 235.313 | 1.05432 | 0.118533 |
| BABA 1h | 1411.55 | 103.489 | 1.15671 | 0.116858 |
| Chitin 1h | 1177.21 | 197.976 | 1.01195 | 0.20667 |
| Epi 1h | 1093.41 | 63.21 | 1.00282 | 0.0579623 |
| SA 1h | 2048.86 | 321.465 | 1.54293 | 0.208181 |
| Me-JA 1h | 2126.99 | 258.091 | 2.03986 | 0.11781 |
| Control 6h | 1421.18 | 259.193 | 1.08415 | 0.113827 |
| ABA 6h | 1325.44 | 214.548 | 0.965831 | 0.0987329 |
| ACC 6h | 1340.92 | 95.8 | 0.92306 | 0.122433 |
| BABA 6h | 1723.67 | 380.362 | 1.16641 | 0.239684 |
| Chitin 6h | 944.883 | 83.4129 | 0.699433 | 0.0587402 |
| Epi 6h | 1025.35 | 121.341 | 0.711647 | 0.125434 |
| SA 6h | 903.635 | 74.1549 | 0.720189 | 0.041667 |
| Me-JA 6h | 1669.73 | 159.043 | 1.31851 | 0.227249 |
Source Transcript PGSC0003DMT400067771 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G27500.1 | +3 | 2e-156 | 455 | 223/330 (68%) | Glycosyl hydrolase superfamily protein | chr2:11752364-11753844 REVERSE LENGTH=392 |
