Probe CUST_41497_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41497_PI426222305 | JHI_St_60k_v1 | DMT400029751 | GAGTTGTAAAGCTGCTGTGAACTCCTCAAAAAGCTCTCTCTTCTCTCTTTCTTTCTTTCT |
All Microarray Probes Designed to Gene DMG400011433
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41504_PI426222305 | JHI_St_60k_v1 | DMT400029748 | CTTGCTTCGGTTTTTTCCTCTTGCTAAATGATATTCAAGGGGTTAAATGAAGTTTGAACA |
| CUST_41548_PI426222305 | JHI_St_60k_v1 | DMT400029749 | GAGTTGTAAAGCTGCTGTGAACTCCTCAAAAAGCTCTCTCTTCTCTCTTTCTTTCTTTCT |
| CUST_41497_PI426222305 | JHI_St_60k_v1 | DMT400029751 | GAGTTGTAAAGCTGCTGTGAACTCCTCAAAAAGCTCTCTCTTCTCTCTTTCTTTCTTTCT |
| CUST_41578_PI426222305 | JHI_St_60k_v1 | DMT400029750 | AGCCTCCATCTGGATCTCTTTTCTCTACACACTATGGGAATCTTTCTTCATCAATATTTG |
Microarray Signals from CUST_41497_PI426222305
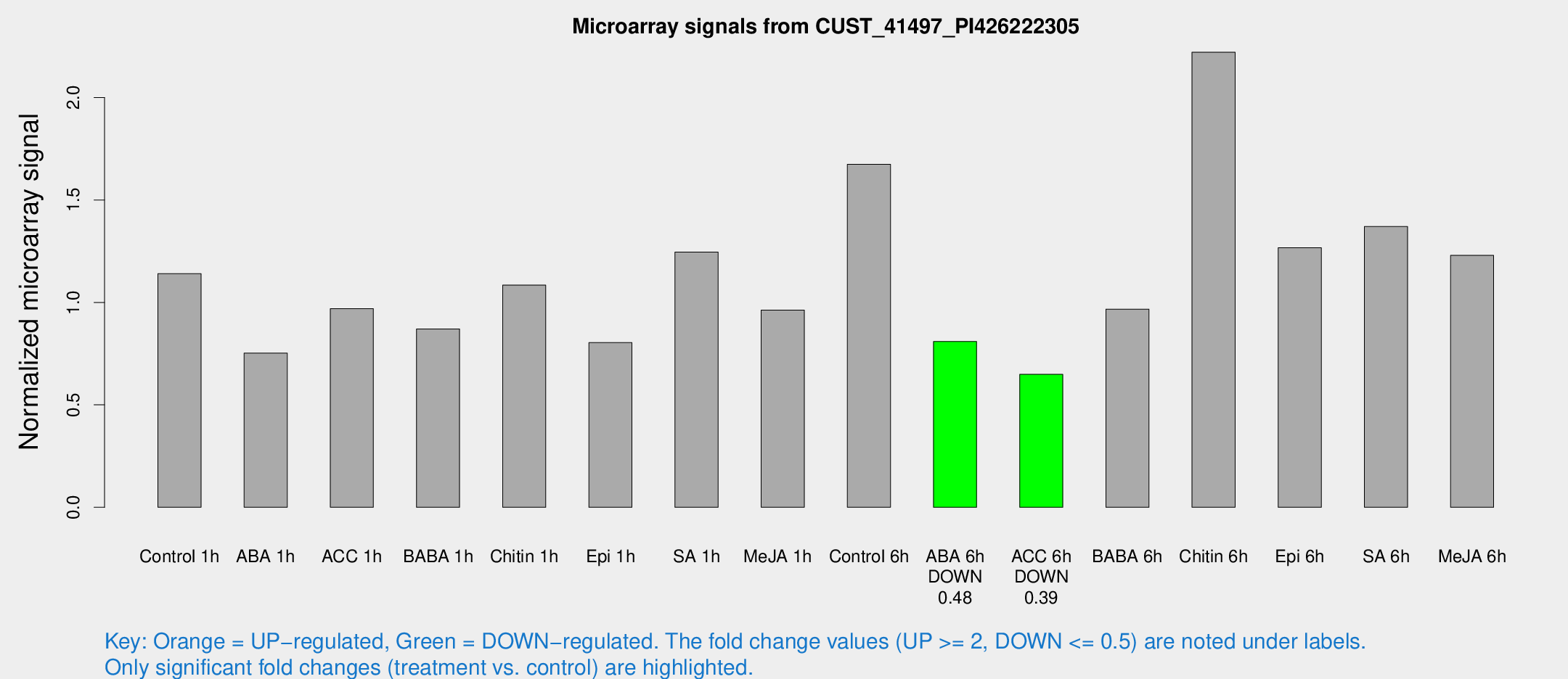
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 79.8577 | 8.61293 | 1.14016 | 0.118413 |
| ABA 1h | 46.6106 | 5.03558 | 0.752397 | 0.110521 |
| ACC 1h | 69.4733 | 6.18153 | 0.969777 | 0.0894308 |
| BABA 1h | 59.0791 | 7.24868 | 0.870476 | 0.0740398 |
| Chitin 1h | 67.6024 | 5.32514 | 1.08439 | 0.08527 |
| Epi 1h | 48.4445 | 4.51132 | 0.803729 | 0.0757988 |
| SA 1h | 90.5031 | 13.0453 | 1.24569 | 0.155937 |
| Me-JA 1h | 54.7737 | 4.65382 | 0.962359 | 0.150458 |
| Control 6h | 116.741 | 7.91691 | 1.6739 | 0.11036 |
| ABA 6h | 59.6102 | 5.09239 | 0.809287 | 0.0690986 |
| ACC 6h | 51.81 | 5.48366 | 0.648806 | 0.0837065 |
| BABA 6h | 77.2786 | 13.0672 | 0.967094 | 0.146048 |
| Chitin 6h | 209.221 | 107.017 | 2.22109 | 1.38185 |
| Epi 6h | 99.2539 | 7.23763 | 1.26678 | 0.0920816 |
| SA 6h | 110.847 | 47.4036 | 1.37032 | 0.867841 |
| Me-JA 6h | 85.6107 | 8.44783 | 1.22935 | 0.128254 |
Source Transcript PGSC0003DMT400029751 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42200.1 | +2 | 1e-52 | 187 | 139/375 (37%) | squamosa promoter binding protein-like 9 | chr2:17587601-17589451 FORWARD LENGTH=375 |
