Probe CUST_41483_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41483_PI426222305 | JHI_St_60k_v1 | DMT400021571 | CACTATAATAGTCAAATTTCCTTTGGAAGCATCCATAAGATATTTGGGTAAATGGCATCG |
All Microarray Probes Designed to Gene DMG400008363
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41581_PI426222305 | JHI_St_60k_v1 | DMT400021570 | GGTCACATTGATGCTCTTATTAACAATGCTGGCGTTAGAGAATTTATCAATTTCGGTTCG |
| CUST_41483_PI426222305 | JHI_St_60k_v1 | DMT400021571 | CACTATAATAGTCAAATTTCCTTTGGAAGCATCCATAAGATATTTGGGTAAATGGCATCG |
Microarray Signals from CUST_41483_PI426222305
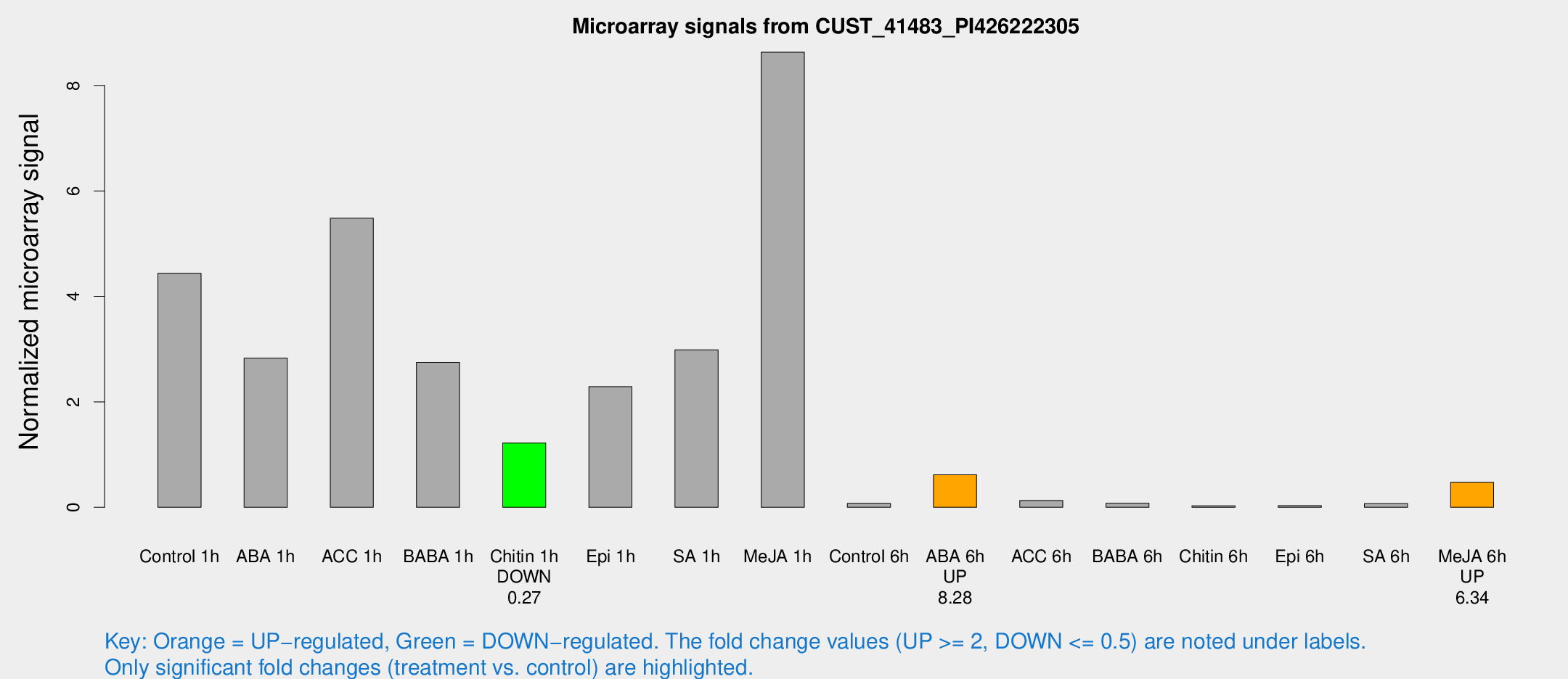
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3231.12 | 771.25 | 4.43903 | 0.737623 |
| ABA 1h | 1998.09 | 631.55 | 2.82696 | 1.14633 |
| ACC 1h | 4567.37 | 1777.61 | 5.48459 | 2.1006 |
| BABA 1h | 1902.61 | 389.696 | 2.75028 | 0.40084 |
| Chitin 1h | 784.47 | 139.424 | 1.21812 | 0.167977 |
| Epi 1h | 1388.13 | 171.288 | 2.28885 | 0.283113 |
| SA 1h | 2538.2 | 1059.46 | 2.98724 | 1.61494 |
| Me-JA 1h | 5016.61 | 925.23 | 8.63053 | 0.860514 |
| Control 6h | 63.6114 | 29.4812 | 0.074452 | 0.0312847 |
| ABA 6h | 476.713 | 108.979 | 0.616825 | 0.127127 |
| ACC 6h | 126.59 | 58.6551 | 0.129955 | 0.082795 |
| BABA 6h | 66.9009 | 22.7862 | 0.0779326 | 0.0241713 |
| Chitin 6h | 25.6954 | 11.1791 | 0.0291879 | 0.0144311 |
| Epi 6h | 30.0936 | 11.1895 | 0.0312384 | 0.0240512 |
| SA 6h | 47.4389 | 4.88375 | 0.0693123 | 0.011012 |
| Me-JA 6h | 364.234 | 130.359 | 0.472208 | 0.220895 |
Source Transcript PGSC0003DMT400021571 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G63380.1 | +1 | 3e-109 | 325 | 166/277 (60%) | NAD(P)-binding Rossmann-fold superfamily protein | chr1:23505582-23506504 FORWARD LENGTH=282 |
