Probe CUST_40338_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40338_PI426222305 | JHI_St_60k_v1 | DMT400048394 | TTGTTATGGTGGCATTGATTGCTCTTTATTTTCCTCCAATTGTTCTGCTAATGTTGAAGG |
All Microarray Probes Designed to Gene DMG401018799
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40338_PI426222305 | JHI_St_60k_v1 | DMT400048394 | TTGTTATGGTGGCATTGATTGCTCTTTATTTTCCTCCAATTGTTCTGCTAATGTTGAAGG |
| CUST_40308_PI426222305 | JHI_St_60k_v1 | DMT400048395 | TCTTCATCTGTGGGTAATCAACATTCTTCATGACCATCATGTTGTGTATTGCAAATCTAG |
| CUST_40313_PI426222305 | JHI_St_60k_v1 | DMT400048390 | TCATTCATCTCTAGTCTCTACCTCACTTGTAGAACTACACCGAGTATATTATTGTTATTG |
Microarray Signals from CUST_40338_PI426222305
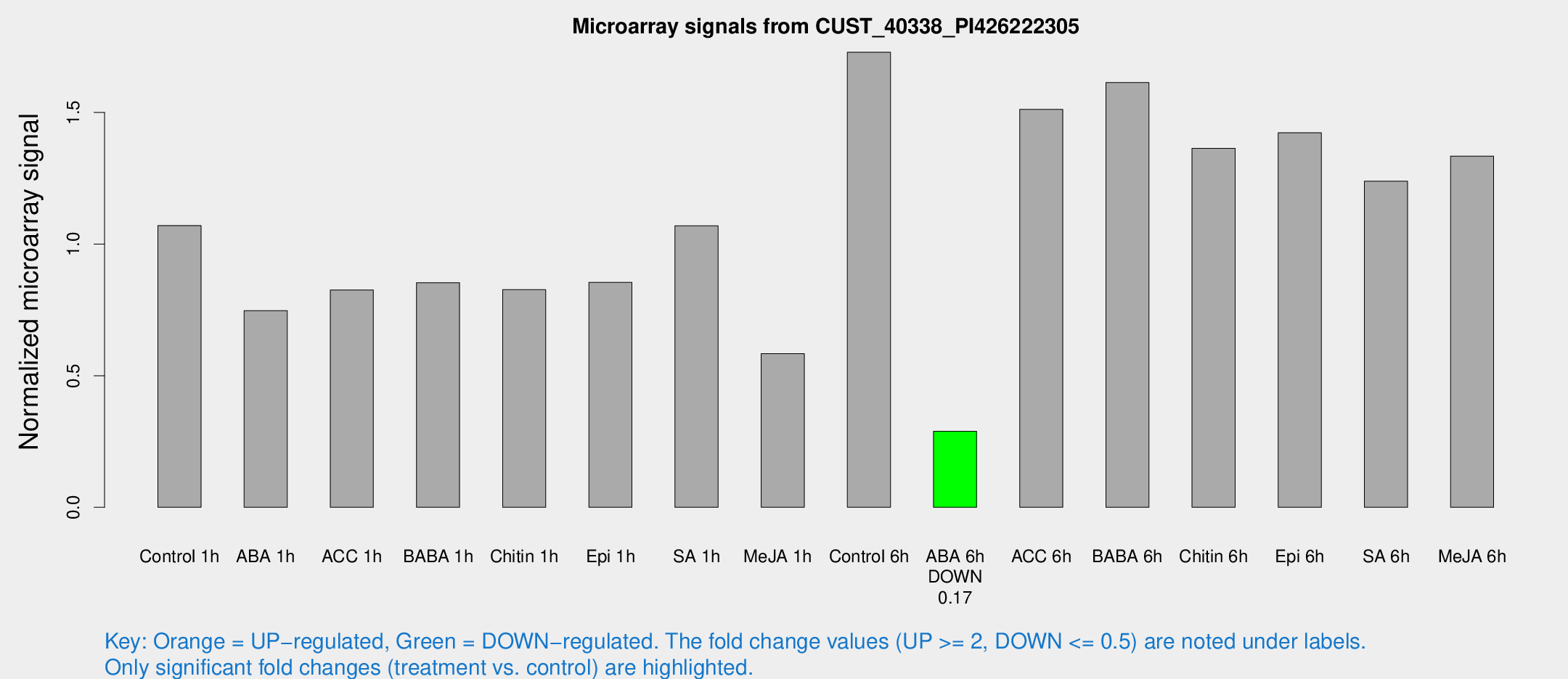
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 59.2893 | 20.1637 | 1.07041 | 0.284233 |
| ABA 1h | 34.1801 | 6.1733 | 0.747101 | 0.0983242 |
| ACC 1h | 48.9003 | 15.7154 | 0.825785 | 0.298768 |
| BABA 1h | 41.3149 | 4.10012 | 0.853285 | 0.0854288 |
| Chitin 1h | 37.486 | 4.00343 | 0.826738 | 0.12287 |
| Epi 1h | 37.4922 | 4.86521 | 0.854667 | 0.0875663 |
| SA 1h | 56.9666 | 11.7212 | 1.06966 | 0.220988 |
| Me-JA 1h | 24.3001 | 3.48786 | 0.583627 | 0.0861482 |
| Control 6h | 97.5938 | 28.5387 | 1.72875 | 0.520472 |
| ABA 6h | 15.3692 | 3.42398 | 0.289176 | 0.064469 |
| ACC 6h | 88.6293 | 14.3797 | 1.51148 | 0.107852 |
| BABA 6h | 91.6619 | 11.4825 | 1.61347 | 0.160841 |
| Chitin 6h | 73.8183 | 9.9022 | 1.36394 | 0.15156 |
| Epi 6h | 84.8547 | 20.4364 | 1.42268 | 0.424296 |
| SA 6h | 61.1601 | 4.89883 | 1.23868 | 0.172624 |
| Me-JA 6h | 70.2578 | 15.4606 | 1.33422 | 0.256738 |
Source Transcript PGSC0003DMT400048394 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G34040.1 | +3 | 4e-94 | 305 | 148/283 (52%) | Pyridoxal phosphate (PLP)-dependent transferases superfamily protein | chr1:12374433-12376179 FORWARD LENGTH=457 |
