Probe CUST_39956_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39956_PI426222305 | JHI_St_60k_v1 | DMT400076999 | CCTAGAATTGTGAAGAAAGTTGGGTTGGAATTACAAGTTCTTTTCGATTATGCGTTTAGT |
All Microarray Probes Designed to Gene DMG400029948
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39956_PI426222305 | JHI_St_60k_v1 | DMT400076999 | CCTAGAATTGTGAAGAAAGTTGGGTTGGAATTACAAGTTCTTTTCGATTATGCGTTTAGT |
Microarray Signals from CUST_39956_PI426222305
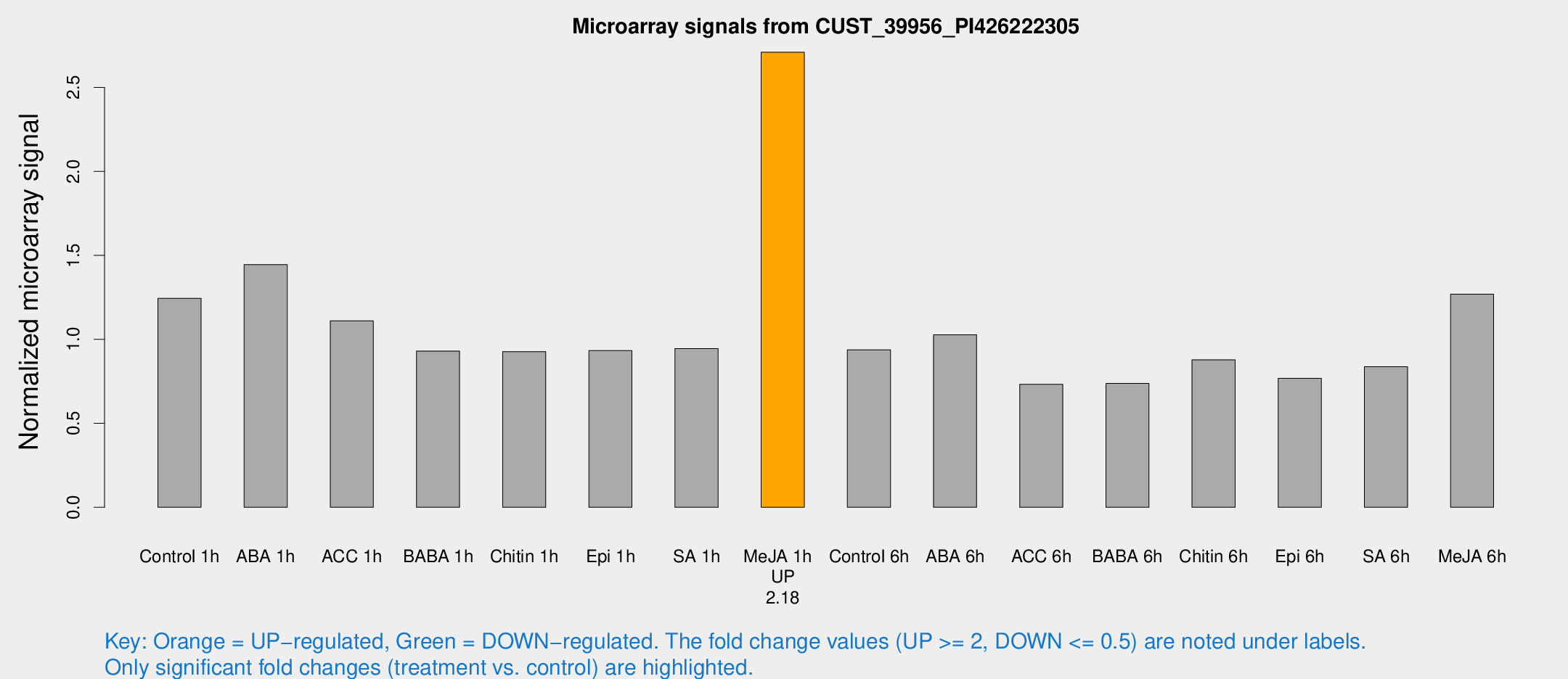
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 656.158 | 127.112 | 1.24425 | 0.156398 |
| ABA 1h | 655.979 | 62.131 | 1.44478 | 0.0879313 |
| ACC 1h | 588.176 | 68.9426 | 1.10981 | 0.0644321 |
| BABA 1h | 460.426 | 44.6475 | 0.930308 | 0.054145 |
| Chitin 1h | 431.87 | 60.0446 | 0.927047 | 0.12019 |
| Epi 1h | 417.569 | 52.2299 | 0.933139 | 0.106285 |
| SA 1h | 503.623 | 67.413 | 0.945401 | 0.198328 |
| Me-JA 1h | 1136.62 | 119.018 | 2.70905 | 0.1566 |
| Control 6h | 492.901 | 84.2241 | 0.937355 | 0.0833072 |
| ABA 6h | 573.468 | 97.7335 | 1.02706 | 0.215543 |
| ACC 6h | 430.798 | 42.2753 | 0.731899 | 0.0427637 |
| BABA 6h | 421.26 | 27.0585 | 0.737367 | 0.0430614 |
| Chitin 6h | 481.024 | 55.105 | 0.878025 | 0.103517 |
| Epi 6h | 454.296 | 77.6922 | 0.767494 | 0.190179 |
| SA 6h | 427.123 | 50.6676 | 0.837243 | 0.0488636 |
| Me-JA 6h | 647.455 | 57.3214 | 1.26837 | 0.0735238 |
Source Transcript PGSC0003DMT400076999 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G11890.1 | +3 | 2e-24 | 102 | 92/239 (38%) | FUNCTIONS IN: molecular_function unknown; LOCATED IN: plasma membrane; EXPRESSED IN: 12 plant structures; EXPRESSED DURING: 6 growth stages; BEST Arabidopsis thaliana protein match is: Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family (TAIR:AT1G17620.1); Has 1807 Blast hits to 1807 proteins in 277 species: Archae - 0; Bacteria - 0; Metazoa - 736; Fungi - 347; Plants - 385; Viruses - 0; Other Eukaryotes - 339 (source: NCBI BLink). | chr5:3831770-3832633 FORWARD LENGTH=287 |
