Probe CUST_39742_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39742_PI426222305 | JHI_St_60k_v1 | DMT400067951 | AGTTCAAGATTGGAGAATATGTGTTGGCGCATTTGGCATCTTGCTCGCAAGAAGAAACAG |
All Microarray Probes Designed to Gene DMG400026428
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39742_PI426222305 | JHI_St_60k_v1 | DMT400067951 | AGTTCAAGATTGGAGAATATGTGTTGGCGCATTTGGCATCTTGCTCGCAAGAAGAAACAG |
Microarray Signals from CUST_39742_PI426222305
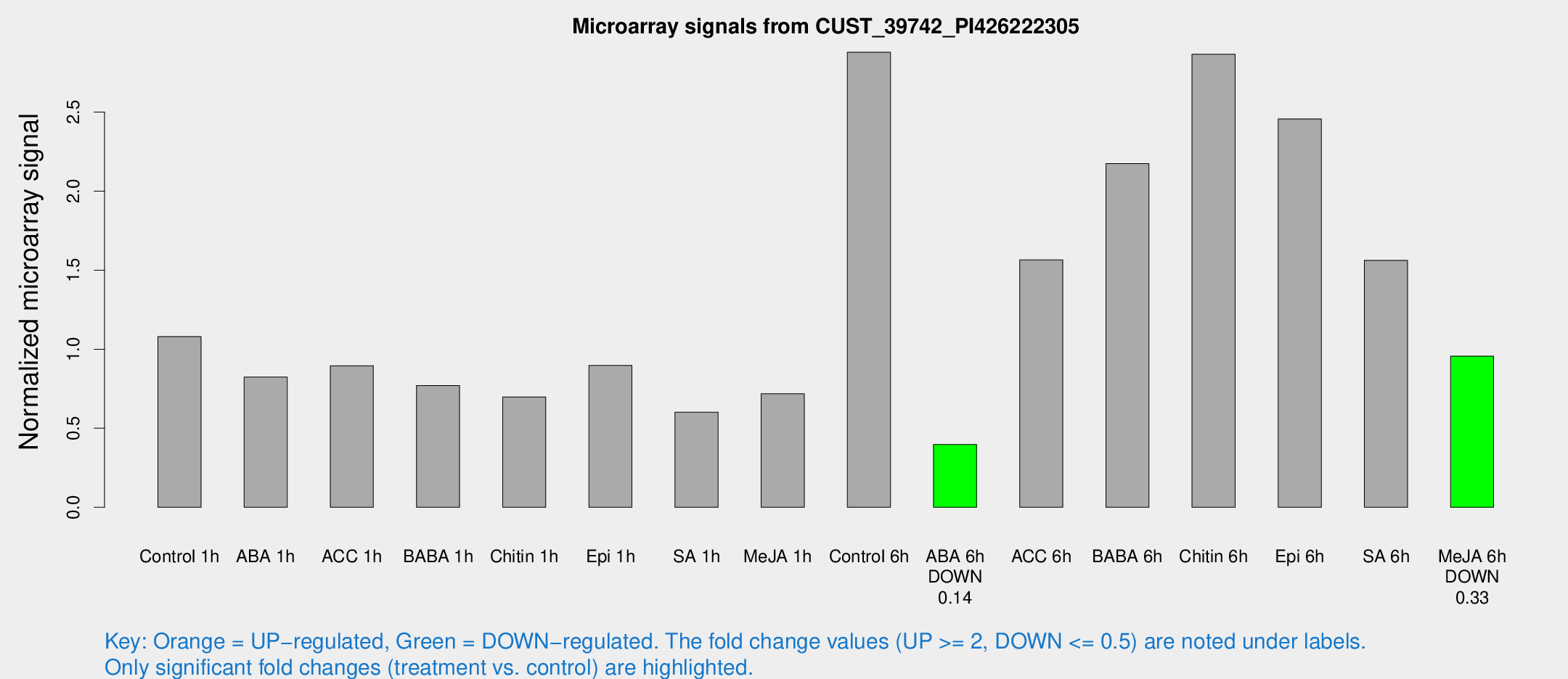
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 134.546 | 11.7345 | 1.08044 | 0.118017 |
| ABA 1h | 92.1818 | 13.4377 | 0.823686 | 0.191715 |
| ACC 1h | 113.86 | 7.63982 | 0.895364 | 0.0834697 |
| BABA 1h | 92.723 | 8.81522 | 0.770713 | 0.0553869 |
| Chitin 1h | 80.3496 | 15.5317 | 0.698076 | 0.153805 |
| Epi 1h | 96.4104 | 6.64968 | 0.897232 | 0.0621512 |
| SA 1h | 78.8295 | 13.859 | 0.601256 | 0.135855 |
| Me-JA 1h | 73.5668 | 9.19694 | 0.718009 | 0.141237 |
| Control 6h | 366.499 | 62.4681 | 2.87879 | 0.40736 |
| ABA 6h | 52.5638 | 4.91807 | 0.396979 | 0.0374778 |
| ACC 6h | 242.789 | 64.6145 | 1.56523 | 0.371506 |
| BABA 6h | 319 | 79.4921 | 2.17421 | 0.499312 |
| Chitin 6h | 389.479 | 64.6858 | 2.86592 | 0.547114 |
| Epi 6h | 361.306 | 77.9745 | 2.45701 | 0.8269 |
| SA 6h | 282.393 | 123.072 | 1.56222 | 2.28363 |
| Me-JA 6h | 131.699 | 45.0389 | 0.956319 | 0.25039 |
Source Transcript PGSC0003DMT400067951 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G04920.1 | +2 | 0.0 | 1552 | 804/1074 (75%) | sucrose phosphate synthase 3F | chr1:1391674-1395756 REVERSE LENGTH=1062 |
