Probe CUST_39495_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39495_PI426222305 | JHI_St_60k_v1 | DMT400019050 | AATTGCTGATTCAAATGCCCCTTCTCTTAATATCGATTTTCGACACGAGTGCATGTTTTG |
All Microarray Probes Designed to Gene DMG400007384
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39499_PI426222305 | JHI_St_60k_v1 | DMT400019052 | AATGCCCCTTCTCTTAATATCGATTTTCGACAGTAAGTAATCATTCATCCTGTCATTCAA |
| CUST_39506_PI426222305 | JHI_St_60k_v1 | DMT400019051 | GCAGCAATTGCTGATTCAAATGCCCCTTCTCTTAATATCGATTTTCGACAAAAGACAACA |
| CUST_39491_PI426222305 | JHI_St_60k_v1 | DMT400019054 | AATGCCCCTTCTCTTAATATCGATTTCCGACAGTAAGTAATCATTCATCCTCTCATTCAA |
| CUST_39495_PI426222305 | JHI_St_60k_v1 | DMT400019050 | AATTGCTGATTCAAATGCCCCTTCTCTTAATATCGATTTTCGACACGAGTGCATGTTTTG |
| CUST_39485_PI426222305 | JHI_St_60k_v1 | DMT400019053 | TTGCACTTTTTACGTACCTGGGTTATCATTTAGTGAATCGATTGTATATTAGCGAGTGCA |
Microarray Signals from CUST_39495_PI426222305
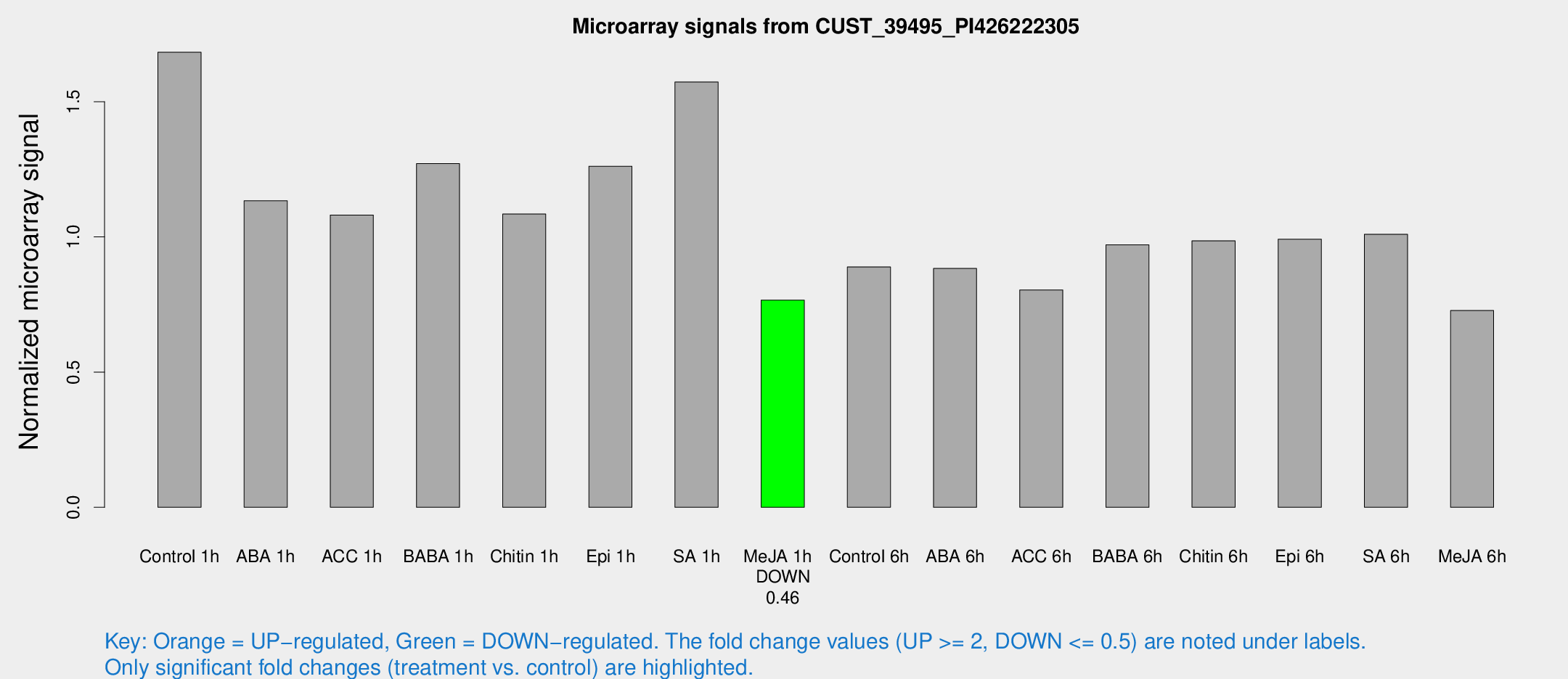
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4617.35 | 267.192 | 1.68284 | 0.0971682 |
| ABA 1h | 2807.44 | 396.165 | 1.13334 | 0.115176 |
| ACC 1h | 3441.35 | 1015.62 | 1.08091 | 0.351795 |
| BABA 1h | 3642.25 | 941.446 | 1.27087 | 0.260189 |
| Chitin 1h | 2670.85 | 154.291 | 1.0845 | 0.067917 |
| Epi 1h | 3005.18 | 216.331 | 1.26127 | 0.0838273 |
| SA 1h | 4439.23 | 257.181 | 1.57283 | 0.0908169 |
| Me-JA 1h | 1717.81 | 100.564 | 0.765917 | 0.0499731 |
| Control 6h | 2525.79 | 468.46 | 0.888858 | 0.115061 |
| ABA 6h | 2614.22 | 328.287 | 0.883152 | 0.0699341 |
| ACC 6h | 2536.63 | 173.991 | 0.803855 | 0.0703466 |
| BABA 6h | 3002.04 | 277.128 | 0.970596 | 0.0560546 |
| Chitin 6h | 2874.49 | 166.125 | 0.985593 | 0.0569224 |
| Epi 6h | 3101.04 | 348.009 | 0.991195 | 0.0572463 |
| SA 6h | 2772.34 | 322.372 | 1.00978 | 0.239918 |
| Me-JA 6h | 2029.41 | 315.011 | 0.727654 | 0.0580223 |
Source Transcript PGSC0003DMT400019050 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G18660.1 | +1 | 2e-10 | 58 | 34/115 (30%) | plant natriuretic peptide A | chr2:8091032-8091644 REVERSE LENGTH=130 |
