Probe CUST_35987_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35987_PI426222305 | JHI_St_60k_v1 | DMT400042828 | TAGCTCACTTAATTTGATGCCTCTTTACTCACAGGTACTAGTTAATGTGTGGACGATTGG |
All Microarray Probes Designed to Gene DMG401016617
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35982_PI426222305 | JHI_St_60k_v1 | DMT400042832 | TGGAGGAAAAGAGAAGATCAGGATTCACAGAAAAGAGTGATGTTCTGGAAATGTTTCTTA |
| CUST_35961_PI426222305 | JHI_St_60k_v1 | DMT400042831 | TACTTCCGCGCAAAGTAGAGCAAGATGTTGAATTGTGTGACTACATCATTCCCAAGGGCT |
| CUST_35987_PI426222305 | JHI_St_60k_v1 | DMT400042828 | TAGCTCACTTAATTTGATGCCTCTTTACTCACAGGTACTAGTTAATGTGTGGACGATTGG |
| CUST_36014_PI426222305 | JHI_St_60k_v1 | DMT400042830 | TGGAGGAAAAGAGAAGATCAGGATTCACAGAAAAGAGTGATGTTCTGGAAATGTTTCTTA |
| CUST_36007_PI426222305 | JHI_St_60k_v1 | DMT400042826 | TTCTTCCTGTATGTCCTCAATGGCGGACGCTTAAGCGAATATTGAACAAAAATATCTTCT |
Microarray Signals from CUST_35987_PI426222305
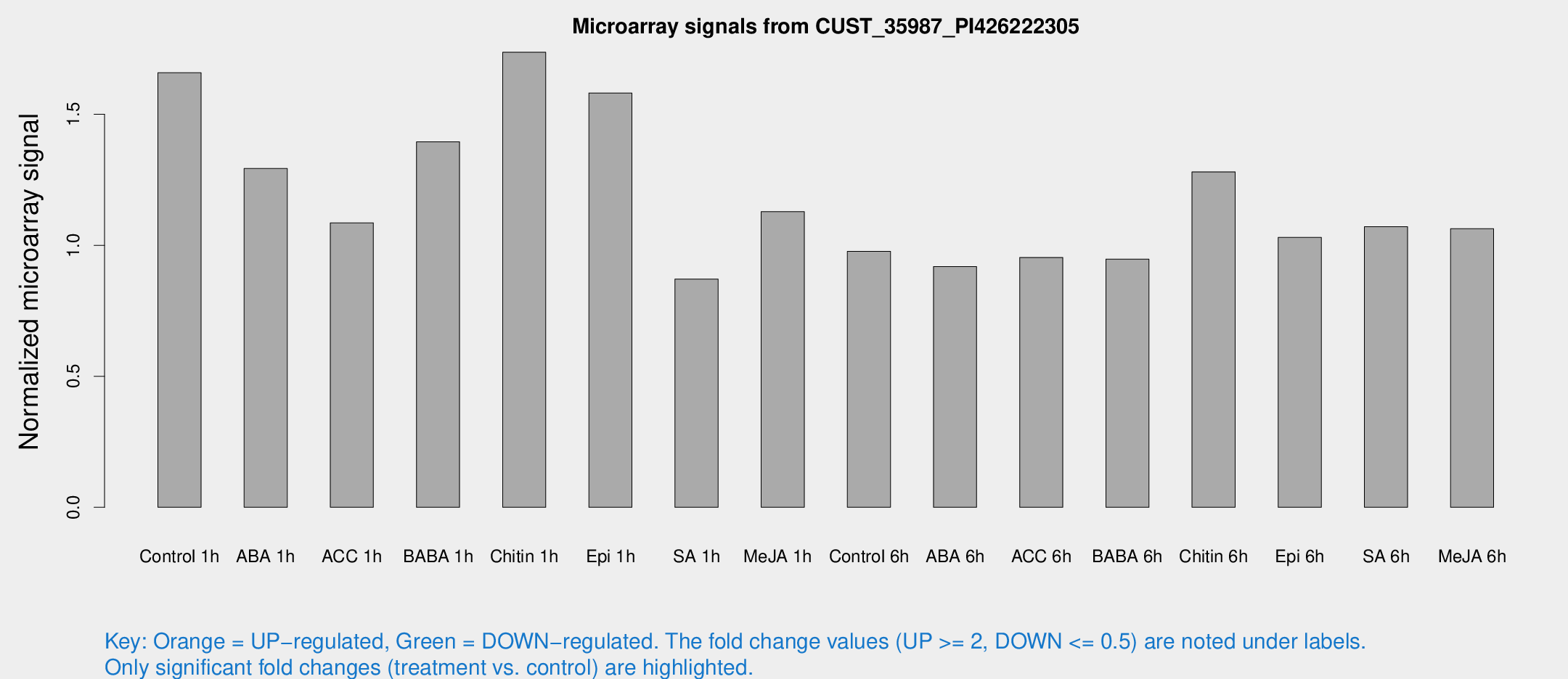
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 10.7181 | 3.23376 | 1.65879 | 0.60812 |
| ABA 1h | 7.15888 | 2.96549 | 1.29308 | 0.620548 |
| ACC 1h | 6.61393 | 3.32151 | 1.08557 | 0.563825 |
| BABA 1h | 9.34676 | 4.09836 | 1.39521 | 0.653656 |
| Chitin 1h | 9.60844 | 3.12956 | 1.73704 | 0.65826 |
| Epi 1h | 8.49074 | 3.04876 | 1.58145 | 0.649262 |
| SA 1h | 5.22589 | 3.03286 | 0.871136 | 0.5052 |
| Me-JA 1h | 5.39255 | 3.13541 | 1.12856 | 0.652807 |
| Control 6h | 5.72099 | 3.2352 | 0.976727 | 0.55299 |
| ABA 6h | 5.7058 | 3.30609 | 0.919159 | 0.53225 |
| ACC 6h | 6.51773 | 3.86009 | 0.953671 | 0.552484 |
| BABA 6h | 6.20549 | 3.60424 | 0.947279 | 0.548551 |
| Chitin 6h | 8.70485 | 3.54699 | 1.28044 | 0.615906 |
| Epi 6h | 6.81084 | 3.66864 | 1.03018 | 0.551747 |
| SA 6h | 6.19769 | 3.37528 | 1.0713 | 0.586858 |
| Me-JA 6h | 6.21904 | 3.23436 | 1.06324 | 0.562092 |
Source Transcript PGSC0003DMT400042828 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G45570.1 | +1 | 2e-39 | 142 | 65/111 (59%) | cytochrome P450, family 76, subfamily C, polypeptide 2 | chr2:18779935-18781922 REVERSE LENGTH=512 |
