Probe CUST_35453_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35453_PI426222305 | JHI_St_60k_v1 | DMT400032522 | GACAAGCTGAAGATGAAACTTAAGCTTCCGGAGCCTTTCAGTTTCATTTTCACATGGTTT |
All Microarray Probes Designed to Gene DMG400012494
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35453_PI426222305 | JHI_St_60k_v1 | DMT400032522 | GACAAGCTGAAGATGAAACTTAAGCTTCCGGAGCCTTTCAGTTTCATTTTCACATGGTTT |
| CUST_35481_PI426222305 | JHI_St_60k_v1 | DMT400032525 | TTCGAGATCACTTATCTTTTGGAGCTTCCGGAGCCTTTCAGTTTCATTTTCACATGGTTT |
| CUST_35451_PI426222305 | JHI_St_60k_v1 | DMT400032523 | CGCATGGAAGAGAGACCATATGATGCAGTTTGTGAAAATGAAAAACAAGTTCAGTATTGA |
| CUST_35530_PI426222305 | JHI_St_60k_v1 | DMT400032526 | TAATATATGTGCTTCCTTCGTATAGAAAACCCGTTCCGTTTACTGAATTCTTGCAAACTG |
Microarray Signals from CUST_35453_PI426222305
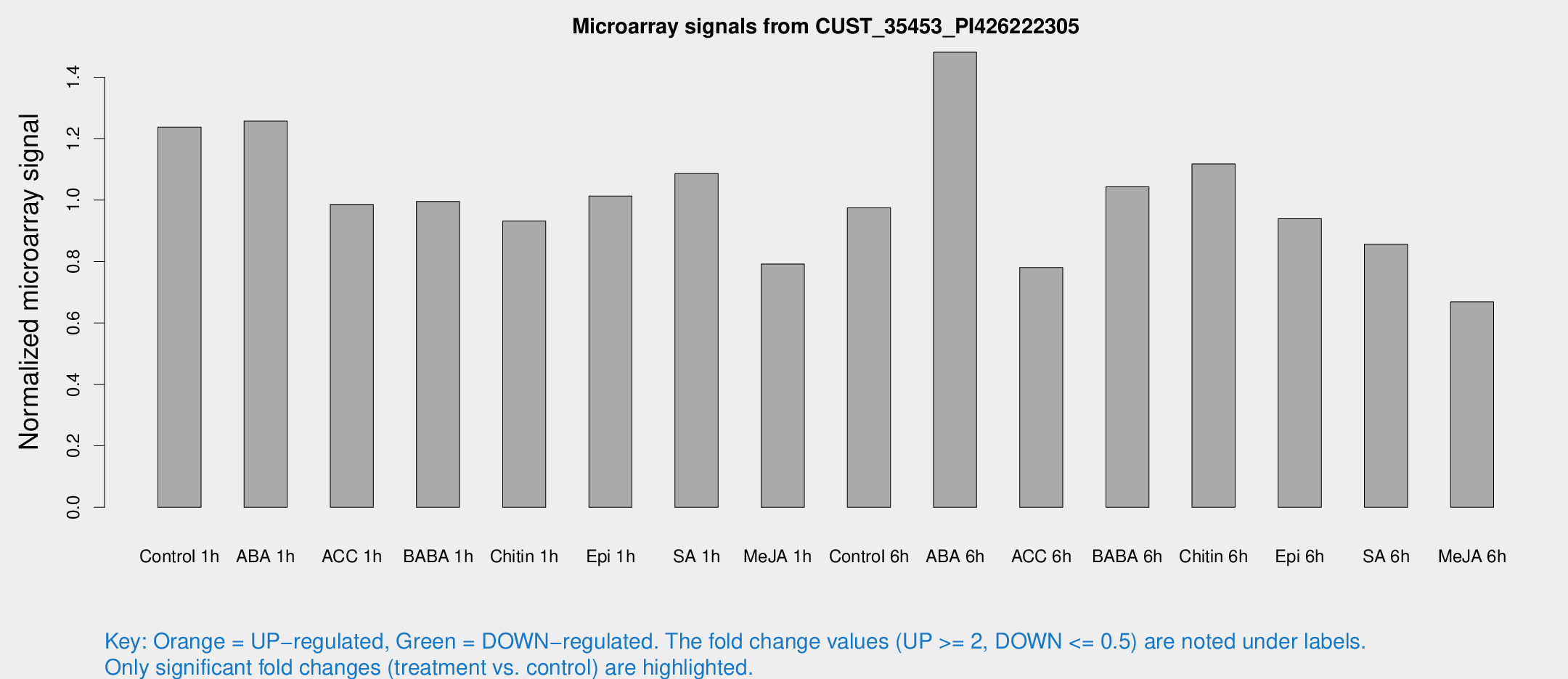
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3846.28 | 318.766 | 1.23765 | 0.0714653 |
| ABA 1h | 3517.71 | 527.738 | 1.25717 | 0.126398 |
| ACC 1h | 3224.52 | 532.817 | 0.986116 | 0.103186 |
| BABA 1h | 3152.17 | 716.05 | 0.995349 | 0.160564 |
| Chitin 1h | 2631.57 | 365.433 | 0.931663 | 0.0760653 |
| Epi 1h | 2726.31 | 244.31 | 1.0129 | 0.0918638 |
| SA 1h | 3556.04 | 661.515 | 1.08658 | 0.13186 |
| Me-JA 1h | 2025.86 | 257.516 | 0.791942 | 0.0457473 |
| Control 6h | 3269.54 | 951.584 | 0.974566 | 0.232947 |
| ABA 6h | 5018.81 | 883.985 | 1.4812 | 0.242066 |
| ACC 6h | 2800.04 | 339.77 | 0.780265 | 0.0450659 |
| BABA 6h | 3676.67 | 538.116 | 1.04307 | 0.134129 |
| Chitin 6h | 3706.9 | 375.482 | 1.11777 | 0.0762365 |
| Epi 6h | 3380.58 | 578.867 | 0.939221 | 0.114484 |
| SA 6h | 2921.49 | 926.014 | 0.856406 | 0.21273 |
| Me-JA 6h | 2160.15 | 439.474 | 0.669076 | 0.103167 |
Source Transcript PGSC0003DMT400032522 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G22890.1 | +1 | 1e-96 | 290 | 171/326 (52%) | PGR5-LIKE A | chr4:12007157-12009175 FORWARD LENGTH=324 |
