Probe CUST_3442_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3442_PI426222305 | JHI_St_60k_v1 | DMT400041206 | TTGGAGATGGAAATCCTTTCCACTGCATCAGTATTTCTTCAATATGCTCTGGATTTATCT |
All Microarray Probes Designed to Gene DMG400015948
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3442_PI426222305 | JHI_St_60k_v1 | DMT400041206 | TTGGAGATGGAAATCCTTTCCACTGCATCAGTATTTCTTCAATATGCTCTGGATTTATCT |
Microarray Signals from CUST_3442_PI426222305
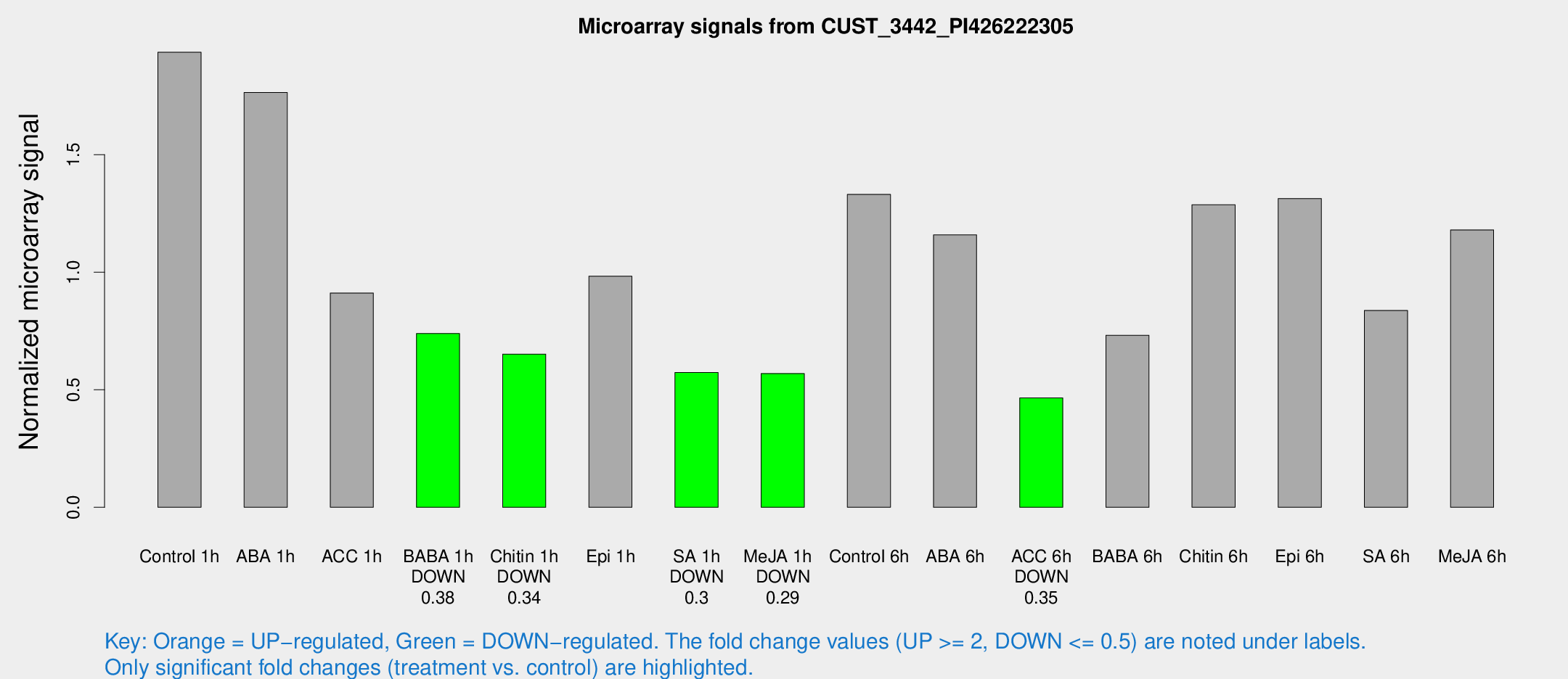
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1062.53 | 103.212 | 1.93565 | 0.186803 |
| ABA 1h | 853.155 | 61.064 | 1.76412 | 0.126506 |
| ACC 1h | 552.8 | 138.224 | 0.911617 | 0.214404 |
| BABA 1h | 415.113 | 100.568 | 0.738988 | 0.130376 |
| Chitin 1h | 328.947 | 58.6823 | 0.651558 | 0.0995696 |
| Epi 1h | 478.269 | 82.7294 | 0.983069 | 0.178649 |
| SA 1h | 326.144 | 44.9817 | 0.573462 | 0.0816348 |
| Me-JA 1h | 253.556 | 15.5278 | 0.568901 | 0.0338837 |
| Control 6h | 732.422 | 69.6897 | 1.33116 | 0.208428 |
| ABA 6h | 670.937 | 38.9857 | 1.15862 | 0.109513 |
| ACC 6h | 296.681 | 44.2612 | 0.465483 | 0.0293812 |
| BABA 6h | 493.133 | 139.097 | 0.731797 | 0.293761 |
| Chitin 6h | 748.085 | 47.3257 | 1.28678 | 0.116891 |
| Epi 6h | 810.051 | 59.5485 | 1.31336 | 0.150982 |
| SA 6h | 484.729 | 120.038 | 0.837317 | 0.146022 |
| Me-JA 6h | 755.768 | 310.21 | 1.18004 | 0.484465 |
Source Transcript PGSC0003DMT400041206 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G19875.1 | +1 | 8e-12 | 60 | 44/86 (51%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: response to oxidative stress; LOCATED IN: endomembrane system; EXPRESSED IN: 21 plant structures; EXPRESSED DURING: 13 growth stages; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT2G31940.1); Has 30201 Blast hits to 17322 proteins in 780 species: Archae - 12; Bacteria - 1396; Metazoa - 17338; Fungi - 3422; Plants - 5037; Viruses - 0; Other Eukaryotes - 2996 (source: NCBI BLink). | chr5:6718412-6718783 FORWARD LENGTH=123 |
