Probe CUST_34191_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34191_PI426222305 | JHI_St_60k_v1 | DMT400030672 | CCTTGCATTGGATCTTAACAATTGATGATGTATGTAATAGGTTTGTGCCAACGAAAACTA |
All Microarray Probes Designed to Gene DMG400011750
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34195_PI426222305 | JHI_St_60k_v1 | DMT400030673 | GCGAAACACAGAAAAATTCTTAATCCGGCTTTCCACATGGAAAAATTGAAGGTATTTACT |
| CUST_34185_PI426222305 | JHI_St_60k_v1 | DMT400030674 | AATAAAATAGCCAAGTCTAAACCCATGCCATTGCACCACGATTTCACACCTCGACTTAAT |
| CUST_34211_PI426222305 | JHI_St_60k_v1 | DMT400030671 | AGGTAACTATGATCTTGCACGAGGTTCTGAGGTTATATCCATCAGGTTCTCTTGTTAGAG |
| CUST_34191_PI426222305 | JHI_St_60k_v1 | DMT400030672 | CCTTGCATTGGATCTTAACAATTGATGATGTATGTAATAGGTTTGTGCCAACGAAAACTA |
Microarray Signals from CUST_34191_PI426222305
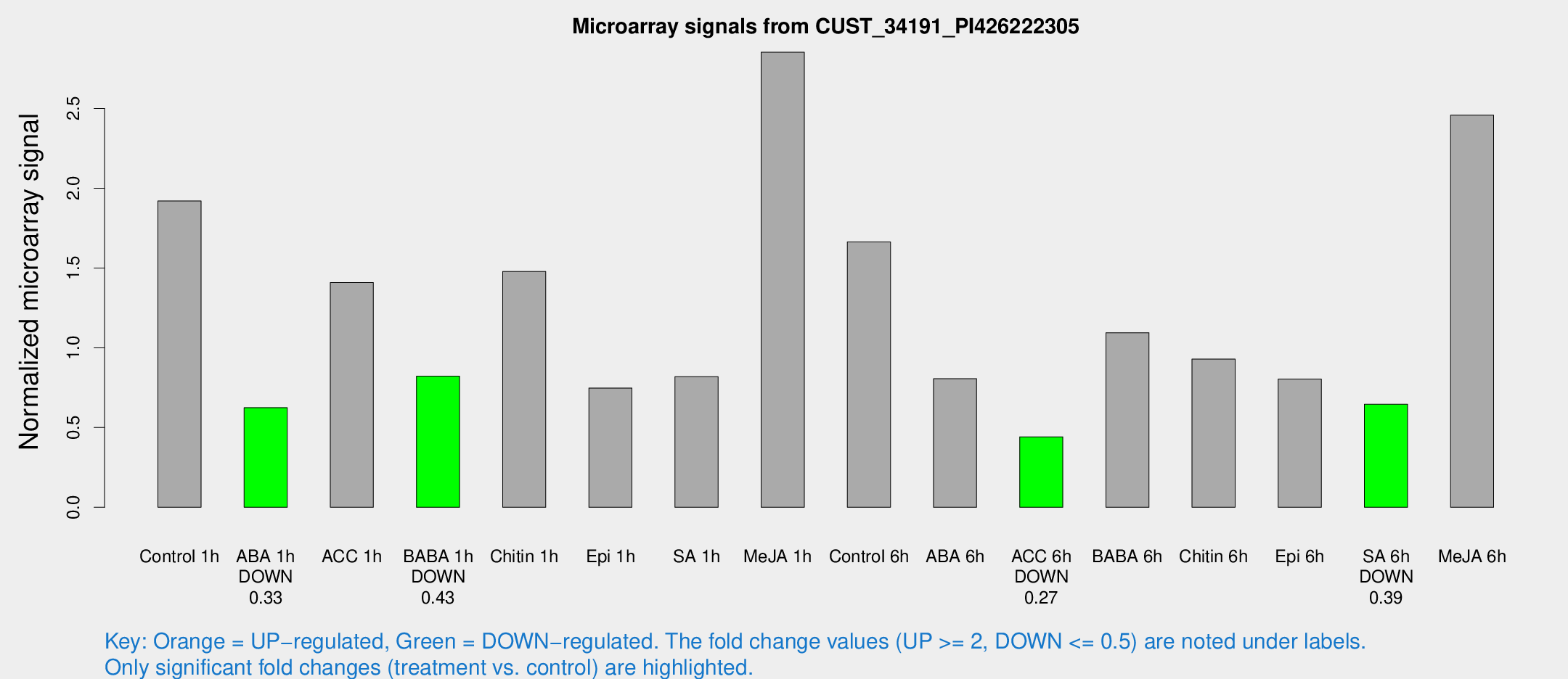
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29.4158 | 4.6912 | 1.92005 | 0.47975 |
| ABA 1h | 8.55649 | 3.54983 | 0.624707 | 0.277206 |
| ACC 1h | 22.3589 | 4.44186 | 1.40862 | 0.292097 |
| BABA 1h | 12.3229 | 3.91986 | 0.822115 | 0.28631 |
| Chitin 1h | 21.6169 | 6.06748 | 1.47814 | 0.4168 |
| Epi 1h | 9.63169 | 3.6086 | 0.747776 | 0.2802 |
| SA 1h | 14.6572 | 5.70534 | 0.819186 | 0.342132 |
| Me-JA 1h | 34.7388 | 4.32803 | 2.85254 | 0.357547 |
| Control 6h | 26.406 | 6.68916 | 1.66404 | 0.290166 |
| ABA 6h | 14.1051 | 4.34988 | 0.806259 | 0.310363 |
| ACC 6h | 7.72768 | 4.59347 | 0.44161 | 0.255723 |
| BABA 6h | 20.8155 | 6.30183 | 1.09445 | 0.54168 |
| Chitin 6h | 17.1268 | 6.38615 | 0.928822 | 0.462876 |
| Epi 6h | 14.2078 | 4.44392 | 0.803553 | 0.296952 |
| SA 6h | 9.69148 | 3.98011 | 0.645944 | 0.279885 |
| Me-JA 6h | 59.0318 | 38.3007 | 2.45817 | 2.39738 |
Source Transcript PGSC0003DMT400030672 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14610.1 | +3 | 1e-110 | 288 | 143/273 (52%) | cytochrome P450, family 72, subfamily A, polypeptide 7 | chr3:4912565-4914503 FORWARD LENGTH=512 |
