Probe CUST_3280_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3280_PI426222305 | JHI_St_60k_v1 | DMT400000219 | ACTAACCAAACCACCTCTCCTCATTATTGTTAACGTAATATGTATGTCTTGCGCAAAAAA |
All Microarray Probes Designed to Gene DMG400000065
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3046_PI426222305 | JHI_St_60k_v1 | DMT400000217 | GTGATTCTAGATGCATTTAGAAGTACAGTAGCTGGATGATGGATATCTGCTAAATTGAGT |
| CUST_3179_PI426222305 | JHI_St_60k_v1 | DMT400000218 | CCTTCCGACCTTTCAATAAATATGTTGAGAATATGTCTCAGGATATTGCAGAAATGAGAA |
| CUST_3280_PI426222305 | JHI_St_60k_v1 | DMT400000219 | ACTAACCAAACCACCTCTCCTCATTATTGTTAACGTAATATGTATGTCTTGCGCAAAAAA |
Microarray Signals from CUST_3280_PI426222305
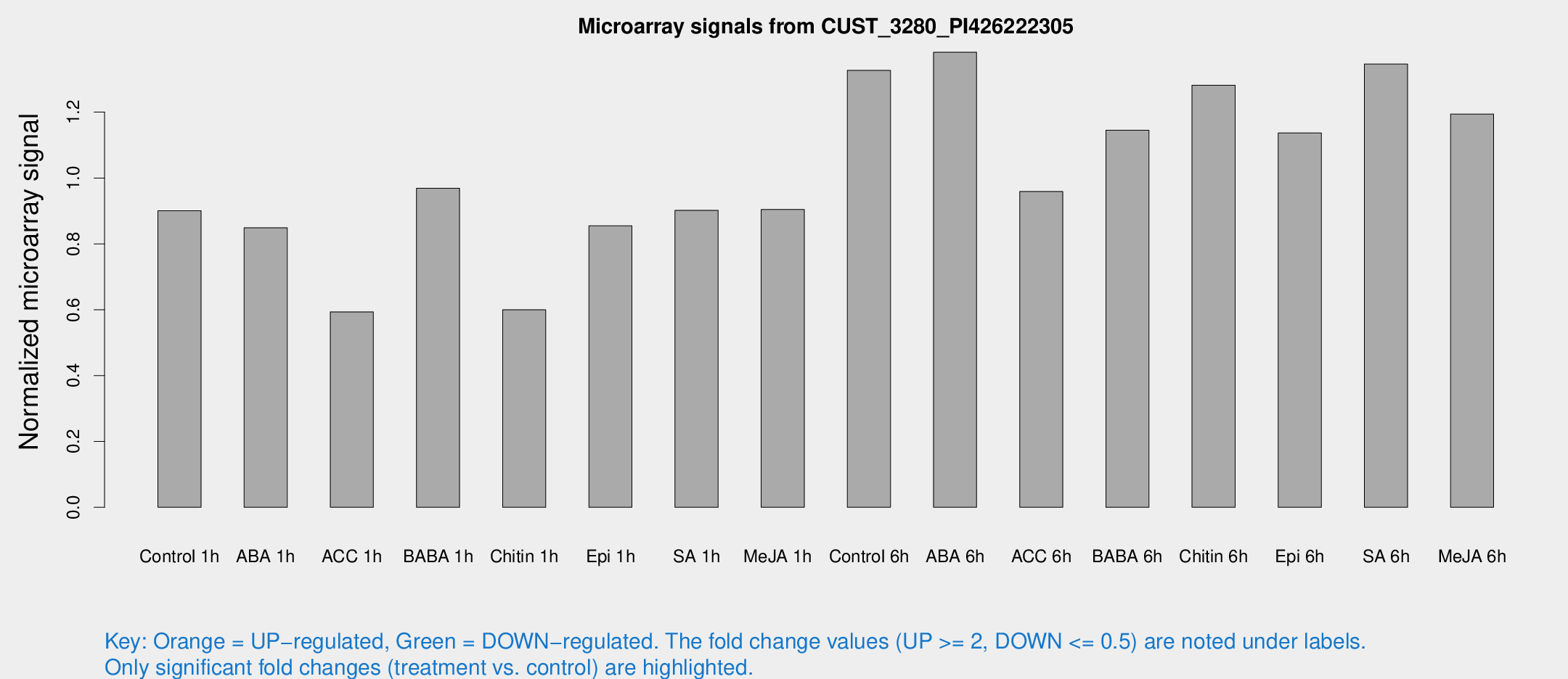
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 93.3271 | 7.98092 | 0.900436 | 0.0595117 |
| ABA 1h | 80.0737 | 15.5078 | 0.84868 | 0.15598 |
| ACC 1h | 70.4775 | 23.0253 | 0.593565 | 0.181536 |
| BABA 1h | 99.8615 | 18.3402 | 0.968869 | 0.0982952 |
| Chitin 1h | 57.8786 | 12.2762 | 0.599799 | 0.0769585 |
| Epi 1h | 78.2333 | 12.4469 | 0.854787 | 0.141045 |
| SA 1h | 97.2283 | 13.0093 | 0.901985 | 0.0841303 |
| Me-JA 1h | 76.8119 | 8.28866 | 0.904512 | 0.0633796 |
| Control 6h | 140.761 | 22.2335 | 1.32686 | 0.106025 |
| ABA 6h | 153.316 | 17.9333 | 1.38195 | 0.0971951 |
| ACC 6h | 115.71 | 17.2284 | 0.959054 | 0.0631128 |
| BABA 6h | 135.713 | 22.5457 | 1.14508 | 0.153299 |
| Chitin 6h | 140.563 | 8.79438 | 1.28176 | 0.0801302 |
| Epi 6h | 132.871 | 11.4554 | 1.13702 | 0.0723048 |
| SA 6h | 145.405 | 37.3941 | 1.34612 | 0.532042 |
| Me-JA 6h | 125.383 | 20.5164 | 1.19411 | 0.108174 |
Source Transcript PGSC0003DMT400000219 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G61540.1 | +2 | 5e-170 | 494 | 274/366 (75%) | alpha/beta-Hydrolases superfamily protein | chr3:22773399-22775699 FORWARD LENGTH=515 |
