Probe CUST_32706_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32706_PI426222305 | JHI_St_60k_v1 | DMT400047323 | ATAAACATGTATAGCGCGTCAAAGGGGAAAAAGAGGAAATTGGAAGTCTTTAATTTGGGC |
All Microarray Probes Designed to Gene DMG400018382
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32726_PI426222305 | JHI_St_60k_v1 | DMT400047325 | ATAAACATGTATAGCGCGTCAAAGGGGAAAAAGAGGAAATTGGAAGTCTTTAATTTGGGC |
| CUST_32706_PI426222305 | JHI_St_60k_v1 | DMT400047323 | ATAAACATGTATAGCGCGTCAAAGGGGAAAAAGAGGAAATTGGAAGTCTTTAATTTGGGC |
| CUST_32767_PI426222305 | JHI_St_60k_v1 | DMT400047324 | ATAAACATGTATAGCGCGTCAAAGGGGAAAAAGAGGAAATTGGAAGTCTTTAATTTGGGC |
Microarray Signals from CUST_32706_PI426222305
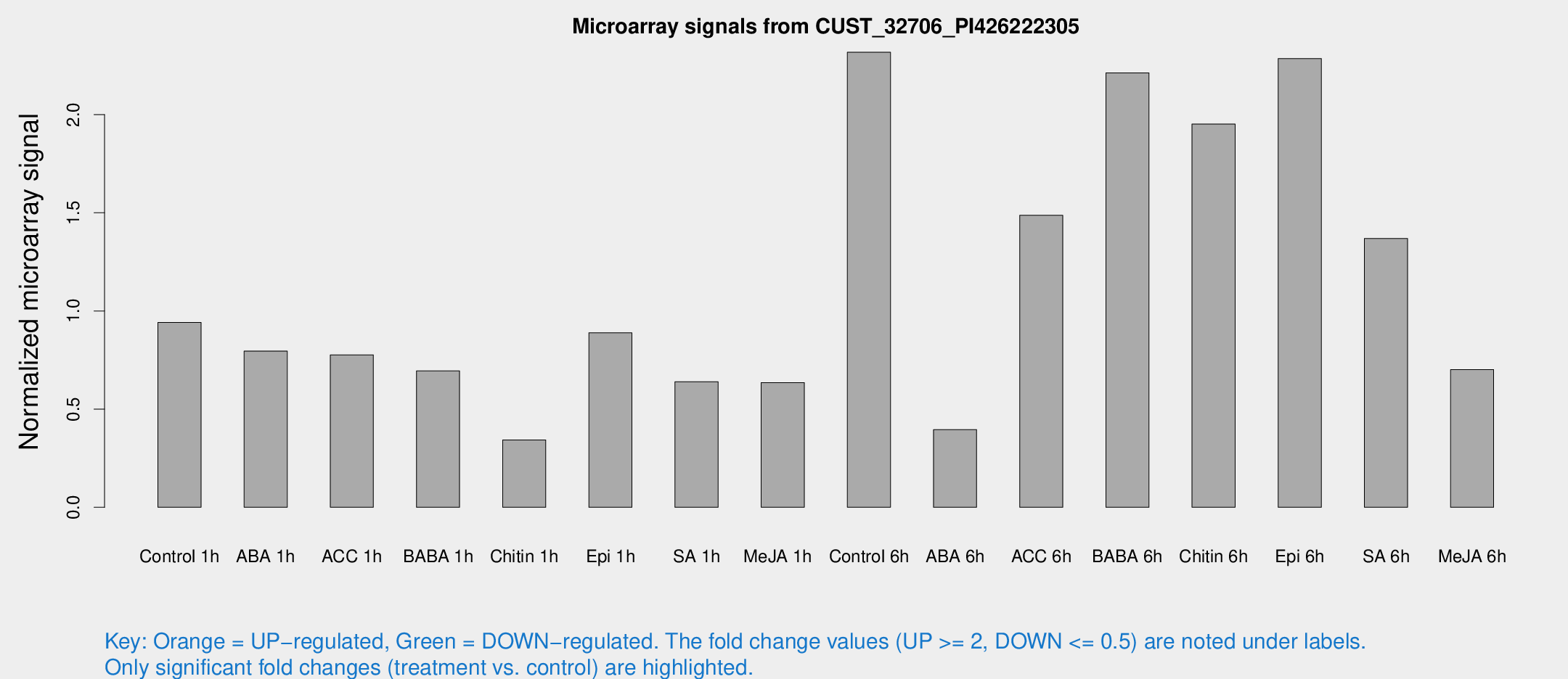
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 32.427 | 3.9781 | 0.941728 | 0.116845 |
| ABA 1h | 25.9099 | 7.54229 | 0.795696 | 0.28531 |
| ACC 1h | 27.8123 | 4.75482 | 0.776294 | 0.173699 |
| BABA 1h | 23.8317 | 4.80247 | 0.694532 | 0.226146 |
| Chitin 1h | 12.251 | 4.54739 | 0.343194 | 0.138609 |
| Epi 1h | 26.3685 | 3.71946 | 0.888891 | 0.127697 |
| SA 1h | 22.5373 | 3.72424 | 0.639807 | 0.106885 |
| Me-JA 1h | 19.6606 | 6.17818 | 0.635303 | 0.304646 |
| Control 6h | 95.7567 | 33.1122 | 2.3178 | 1.01623 |
| ABA 6h | 16.4353 | 5.05035 | 0.395742 | 0.183013 |
| ACC 6h | 59.4814 | 9.11284 | 1.48736 | 0.135559 |
| BABA 6h | 86.2767 | 12.4468 | 2.21293 | 0.237191 |
| Chitin 6h | 75.8069 | 18.8743 | 1.9517 | 0.510992 |
| Epi 6h | 96.0193 | 24.87 | 2.28509 | 0.96868 |
| SA 6h | 49.4324 | 12.7374 | 1.36881 | 0.566326 |
| Me-JA 6h | 29.6473 | 10.6814 | 0.70137 | 0.370025 |
Source Transcript PGSC0003DMT400047323 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G56960.1 | +2 | 1e-54 | 156 | 74/95 (78%) | phosphatidyl inositol monophosphate 5 kinase 4 | chr3:21080957-21083885 FORWARD LENGTH=779 |
