Probe CUST_32171_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32171_PI426222305 | JHI_St_60k_v1 | DMT400037585 | GAAATGGCGTCAACTTCCAATATCAATATCACAGGAAACTTCCATTAATCCACAAGGAGT |
All Microarray Probes Designed to Gene DMG400014497
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32171_PI426222305 | JHI_St_60k_v1 | DMT400037585 | GAAATGGCGTCAACTTCCAATATCAATATCACAGGAAACTTCCATTAATCCACAAGGAGT |
| CUST_32152_PI426222305 | JHI_St_60k_v1 | DMT400037588 | AACTACTTACAGCTTCAGCTTAATTAGAGTGTGGCTTGTGCTCTTTCCATTACATTCGGG |
| CUST_32216_PI426222305 | JHI_St_60k_v1 | DMT400037589 | GGTGGTGAACCTTTGTGGCCAATTTTTAAGTGATCTTTTCCAAATTCCATCTGTTATCTG |
Microarray Signals from CUST_32171_PI426222305
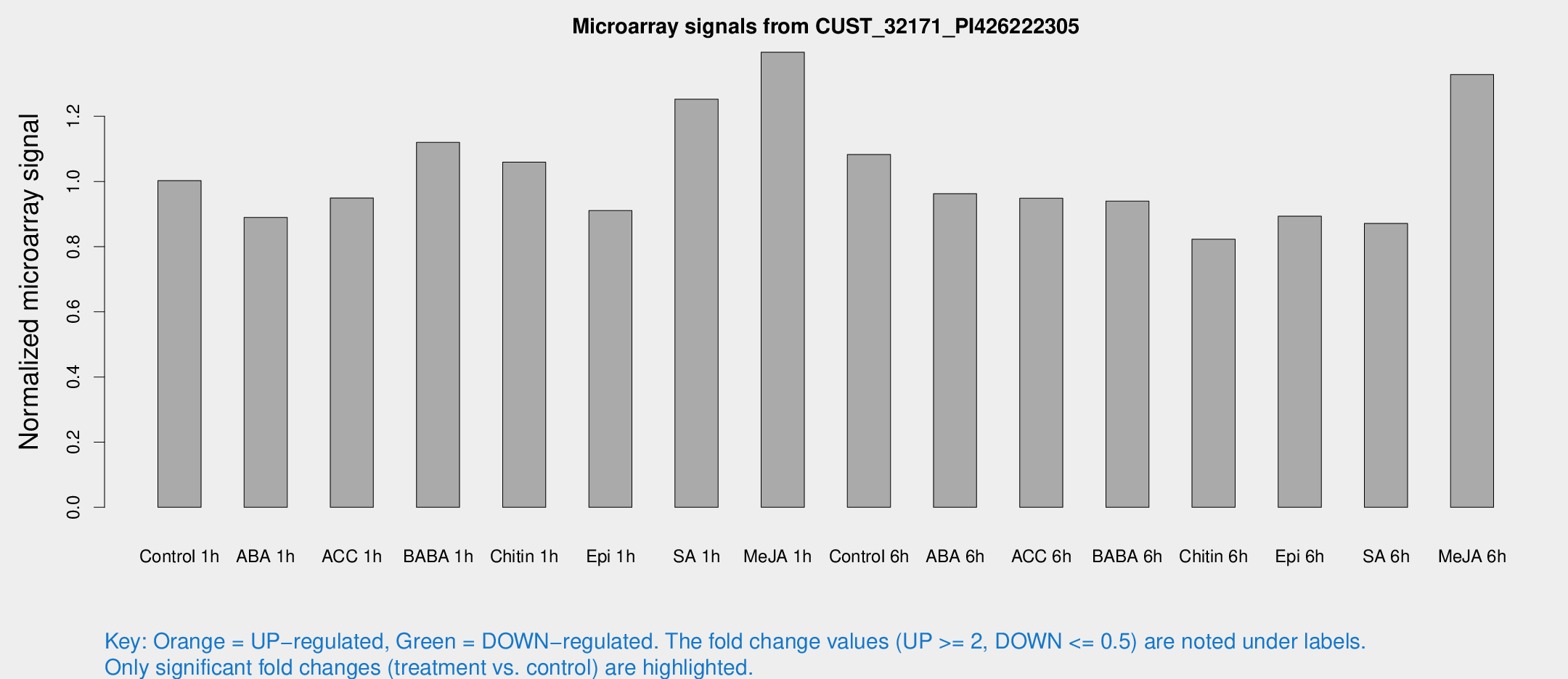
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 619.792 | 97.1376 | 1.00249 | 0.0877955 |
| ABA 1h | 485.728 | 73.7481 | 0.889389 | 0.0704083 |
| ACC 1h | 616.198 | 120.694 | 0.949344 | 0.140814 |
| BABA 1h | 691.15 | 155.132 | 1.1199 | 0.170433 |
| Chitin 1h | 581.753 | 60.5174 | 1.05913 | 0.0615404 |
| Epi 1h | 476.439 | 27.7747 | 0.910496 | 0.0530416 |
| SA 1h | 807.456 | 157.963 | 1.25218 | 0.223135 |
| Me-JA 1h | 706.946 | 117.003 | 1.39657 | 0.107598 |
| Control 6h | 733.738 | 222.998 | 1.08258 | 0.308715 |
| ABA 6h | 621.819 | 47.3027 | 0.962472 | 0.0559123 |
| ACC 6h | 663.629 | 67.1632 | 0.948294 | 0.0591341 |
| BABA 6h | 638.445 | 43.781 | 0.939632 | 0.0545992 |
| Chitin 6h | 529.913 | 30.927 | 0.822942 | 0.0479676 |
| Epi 6h | 621.141 | 92.5968 | 0.893231 | 0.129279 |
| SA 6h | 559.087 | 140.776 | 0.871143 | 0.14836 |
| Me-JA 6h | 817.486 | 124.016 | 1.32811 | 0.167999 |
Source Transcript PGSC0003DMT400037585 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G11490.2 | +3 | 0.0 | 1313 | 660/838 (79%) | adaptin family protein | chr5:3671964-3676263 FORWARD LENGTH=850 |
