Probe CUST_317_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_317_PI426222305 | JHI_St_60k_v1 | DMT400033609 | TGGATAGGCCAAAGAAGGTCTCTTGATGAAATGCTATATCTTGGACGACTGAGATGTCGA |
All Microarray Probes Designed to Gene DMG401012908
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_317_PI426222305 | JHI_St_60k_v1 | DMT400033609 | TGGATAGGCCAAAGAAGGTCTCTTGATGAAATGCTATATCTTGGACGACTGAGATGTCGA |
Microarray Signals from CUST_317_PI426222305
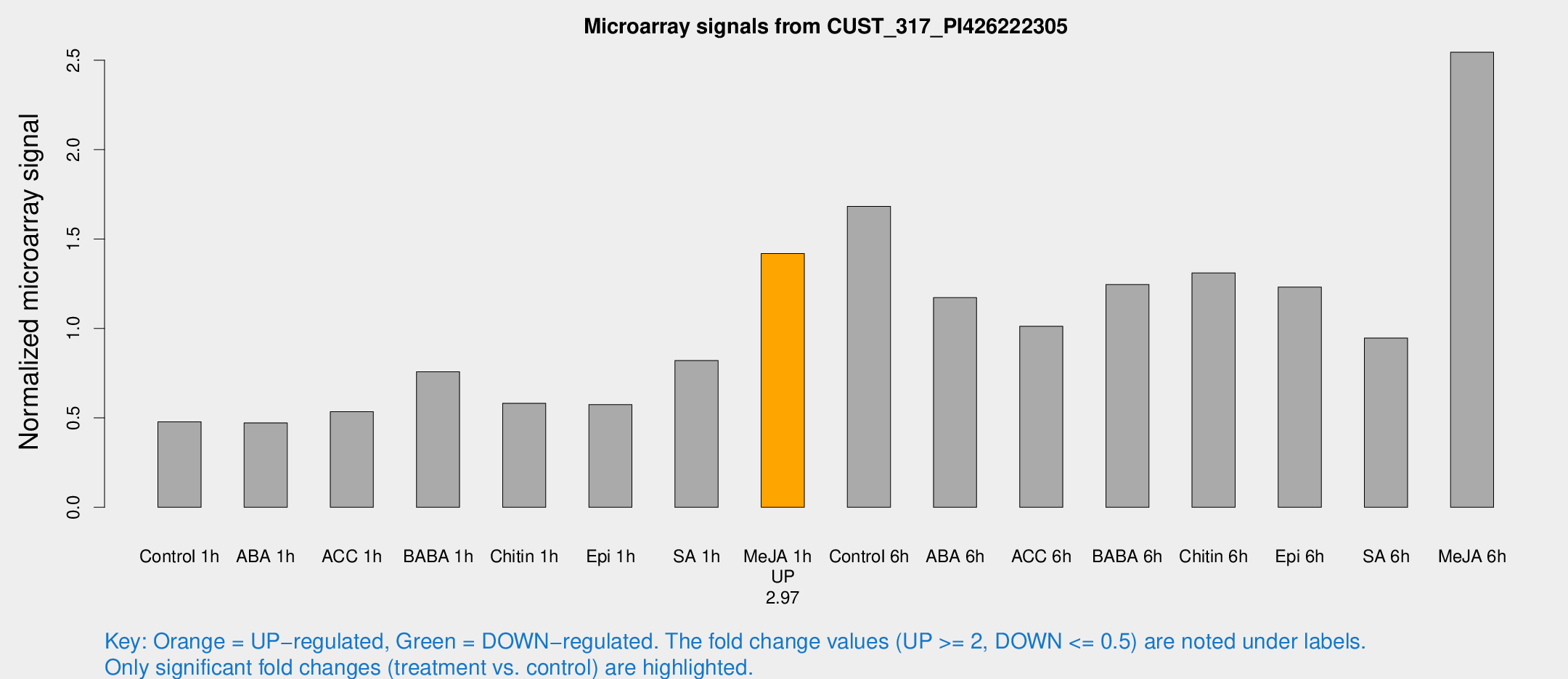
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 126.851 | 19.8452 | 0.478461 | 0.0799025 |
| ABA 1h | 118.875 | 35.9929 | 0.472027 | 0.12346 |
| ACC 1h | 170.747 | 60.6546 | 0.534876 | 0.240075 |
| BABA 1h | 191.165 | 21.3539 | 0.758182 | 0.0458894 |
| Chitin 1h | 136.139 | 12.8935 | 0.581738 | 0.0363049 |
| Epi 1h | 133.591 | 24.7318 | 0.574269 | 0.115882 |
| SA 1h | 246.577 | 91.6008 | 0.820374 | 0.303466 |
| Me-JA 1h | 304.357 | 40.0299 | 1.41944 | 0.0834732 |
| Control 6h | 493.175 | 154.627 | 1.68277 | 0.498393 |
| ABA 6h | 324.746 | 32.9856 | 1.1722 | 0.0688309 |
| ACC 6h | 312.736 | 64.0605 | 1.01258 | 0.130551 |
| BABA 6h | 360.67 | 21.1798 | 1.24615 | 0.0730435 |
| Chitin 6h | 362.903 | 31.0822 | 1.31098 | 0.15176 |
| Epi 6h | 391.994 | 106.364 | 1.2313 | 0.502225 |
| SA 6h | 255.712 | 55.2052 | 0.9464 | 0.126839 |
| Me-JA 6h | 683.092 | 139.163 | 2.54476 | 0.324523 |
Source Transcript PGSC0003DMT400033609 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc04g076120.2 | +1 | 0.0 | 629 | 319/436 (73%) | genomic_reference:SL2.50ch04 gene_region:58665257-58669513 transcript_region:SL2.50ch04:58665257..58669513+ go_terms:GO:0016754 functional_description:Serine carboxypeptidase 1 (AHRD V1 ***- B6TC80_MAIZE); contains Interpro domain(s) IPR001563 Peptidase S10, serine carboxypeptidase |
| TAIR PP10 | AT1G73300.1 | +1 | 7e-133 | 394 | 212/437 (49%) | serine carboxypeptidase-like 2 | chr1:27559673-27562048 REVERSE LENGTH=441 |
