Probe CUST_31013_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31013_PI426222305 | JHI_St_60k_v1 | DMT400040329 | AATCGCGGAGTCTAGTGCAGAAAAGTTCATATGCATCAGCGAGGAGATGGAAGTAGATTG |
All Microarray Probes Designed to Gene DMG400015612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30984_PI426222305 | JHI_St_60k_v1 | DMT400040330 | AATATGCTATCCATTTCAAGGCTGAGGAATCAGCGAGGAGATGGAAGTAGATTGGGCTCT |
| CUST_30954_PI426222305 | JHI_St_60k_v1 | DMT400040331 | TTCAAGGCTGAGGAAGATTGCAACCCCTTTGTAGGAGAAATTTCTATGCTTTCACACACA |
| CUST_31013_PI426222305 | JHI_St_60k_v1 | DMT400040329 | AATCGCGGAGTCTAGTGCAGAAAAGTTCATATGCATCAGCGAGGAGATGGAAGTAGATTG |
| CUST_30977_PI426222305 | JHI_St_60k_v1 | DMT400040328 | GAAAAGTTCATATGCAGATTGCAACCCCTTTGTAGGAGAAATTTCTATGCTTTCACACAC |
Microarray Signals from CUST_31013_PI426222305
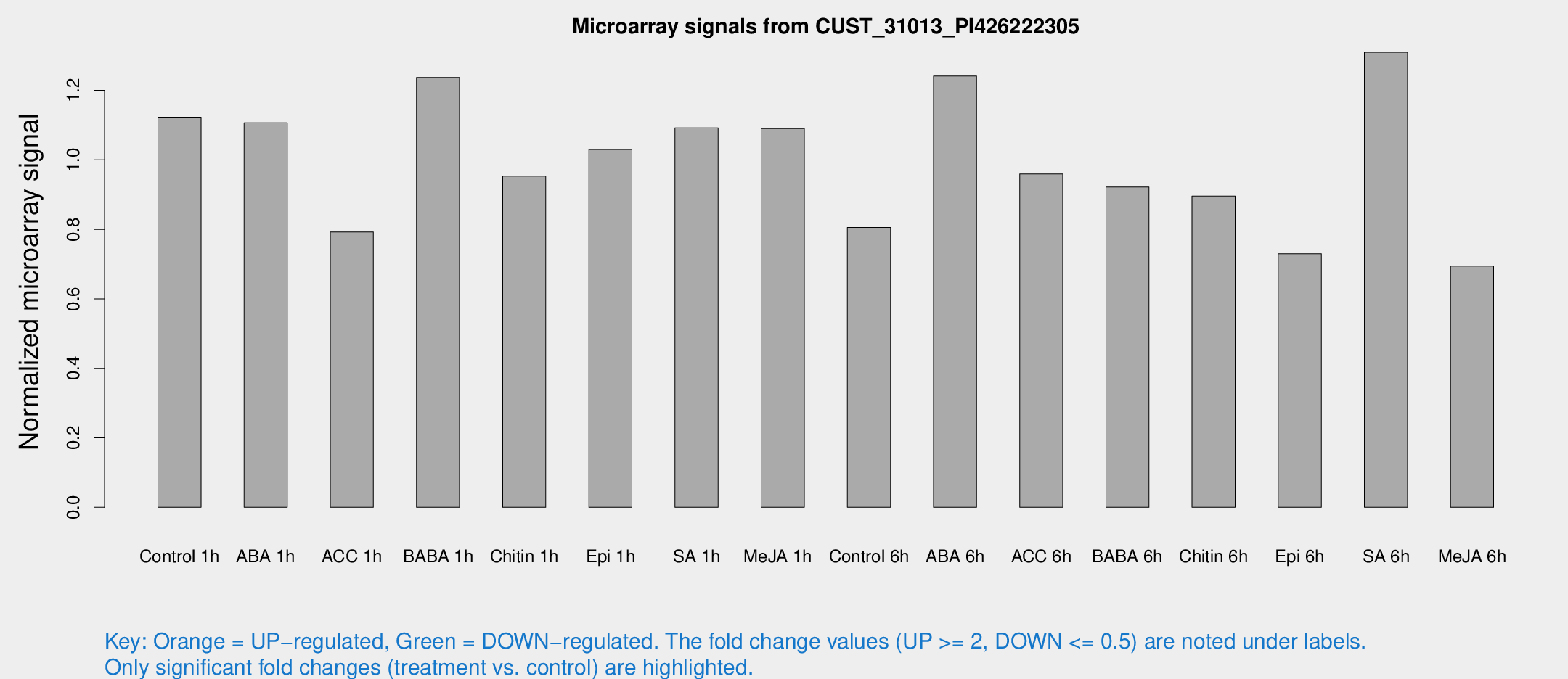
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 90.1683 | 17.2506 | 1.12295 | 0.220424 |
| ABA 1h | 75.9907 | 6.02148 | 1.10669 | 0.0831132 |
| ACC 1h | 67.5737 | 16.2678 | 0.792681 | 0.168389 |
| BABA 1h | 92.5421 | 7.00699 | 1.23706 | 0.0890304 |
| Chitin 1h | 67.086 | 7.70629 | 0.953181 | 0.0778598 |
| Epi 1h | 68.9186 | 5.42155 | 1.03015 | 0.0813423 |
| SA 1h | 86.5671 | 6.21141 | 1.09192 | 0.0783747 |
| Me-JA 1h | 69.3975 | 7.41917 | 1.08998 | 0.198682 |
| Control 6h | 67.8442 | 20.6264 | 0.805448 | 0.205864 |
| ABA 6h | 102.264 | 7.10814 | 1.24119 | 0.0861131 |
| ACC 6h | 86.2113 | 10.1609 | 0.959427 | 0.0986568 |
| BABA 6h | 79.6844 | 6.22215 | 0.921851 | 0.0718423 |
| Chitin 6h | 76.8625 | 16.8533 | 0.89581 | 0.194243 |
| Epi 6h | 65.9095 | 13.2458 | 0.729795 | 0.159251 |
| SA 6h | 103.629 | 19.1226 | 1.30979 | 0.322713 |
| Me-JA 6h | 60.5591 | 20.4925 | 0.694639 | 0.237721 |
Source Transcript PGSC0003DMT400040329 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G70180.2 | +2 | 1e-40 | 154 | 153/476 (32%) | Sterile alpha motif (SAM) domain-containing protein | chr1:26426768-26429097 FORWARD LENGTH=460 |
