Probe CUST_29006_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29006_PI426222305 | JHI_St_60k_v1 | DMT400020689 | GACACACAGCTTGTAGATACAAACCTGAAGAATGTTTTGGTATGTTCAAAAGATGGATAA |
All Microarray Probes Designed to Gene DMG400008018
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29006_PI426222305 | JHI_St_60k_v1 | DMT400020689 | GACACACAGCTTGTAGATACAAACCTGAAGAATGTTTTGGTATGTTCAAAAGATGGATAA |
| CUST_29025_PI426222305 | JHI_St_60k_v1 | DMT400020686 | AACTACTCCATTGTTGATGATTGGCGTCCATGGAACGTAAATAATCAAGTTGCTGGGTAA |
| CUST_29092_PI426222305 | JHI_St_60k_v1 | DMT400020688 | GACACACAGCTTGTAGATACAAACCTGAAGAATGTTTTGGTATGTTCAAAAGATGGATAA |
Microarray Signals from CUST_29006_PI426222305
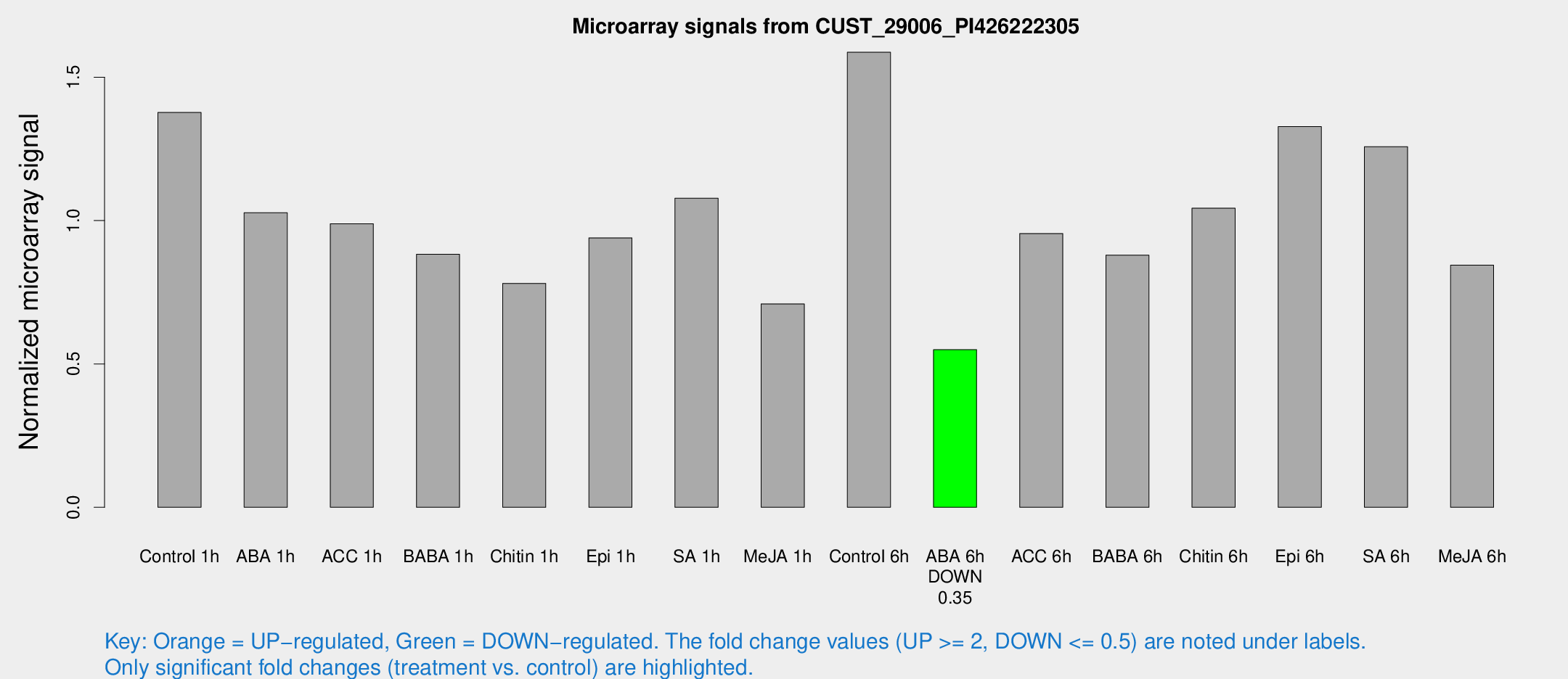
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4379 | 632.45 | 1.37708 | 0.122816 |
| ABA 1h | 2845.97 | 176.455 | 1.02746 | 0.101907 |
| ACC 1h | 3515.71 | 978.03 | 0.988926 | 0.277483 |
| BABA 1h | 2749.02 | 488.592 | 0.882359 | 0.0829603 |
| Chitin 1h | 2193.08 | 126.853 | 0.780875 | 0.0451007 |
| Epi 1h | 2581.65 | 326.08 | 0.939657 | 0.11745 |
| SA 1h | 3493.09 | 364.232 | 1.07804 | 0.0883946 |
| Me-JA 1h | 1808.86 | 104.636 | 0.709495 | 0.0694043 |
| Control 6h | 5192.62 | 1041.13 | 1.58711 | 0.222574 |
| ABA 6h | 1826.14 | 105.677 | 0.549828 | 0.0317644 |
| ACC 6h | 3440.86 | 290.043 | 0.954774 | 0.0698079 |
| BABA 6h | 3198.5 | 659.448 | 0.879455 | 0.190863 |
| Chitin 6h | 3471.32 | 200.906 | 1.04333 | 0.0603215 |
| Epi 6h | 4684.07 | 271.311 | 1.328 | 0.124081 |
| SA 6h | 4102.52 | 986.822 | 1.25774 | 0.201036 |
| Me-JA 6h | 2791.39 | 716.192 | 0.844575 | 0.144096 |
Source Transcript PGSC0003DMT400020689 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G22920.2 | +3 | 1e-150 | 442 | 220/446 (49%) | serine carboxypeptidase-like 12 | chr2:9753938-9757420 FORWARD LENGTH=435 |
