Probe CUST_28563_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28563_PI426222305 | JHI_St_60k_v1 | DMT400055510 | GGATTACAGGAAATTTTCGATCTTGTGTTGACGGTTGCTAAAGAGAGGATATCATTTTTT |
All Microarray Probes Designed to Gene DMG400021553
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28563_PI426222305 | JHI_St_60k_v1 | DMT400055510 | GGATTACAGGAAATTTTCGATCTTGTGTTGACGGTTGCTAAAGAGAGGATATCATTTTTT |
| CUST_28565_PI426222305 | JHI_St_60k_v1 | DMT400055506 | ACTTTCCAGGAAATTTTCGATCTTGTGTTGACGGTTGCTAAAGAGAGGATATCATTTTTT |
| CUST_28493_PI426222305 | JHI_St_60k_v1 | DMT400055509 | GGCGTAGACACCCAAAAAAGAAGTAATGGGATAAATGTTTAGCTGTTTTTAGTTTTGCTT |
| CUST_28520_PI426222305 | JHI_St_60k_v1 | DMT400055507 | GGCGTAGACACCCAAAAAAGAAGTAATGGGATAAATGTTTAGCTGTTTTTAGTTTTGCTT |
Microarray Signals from CUST_28563_PI426222305
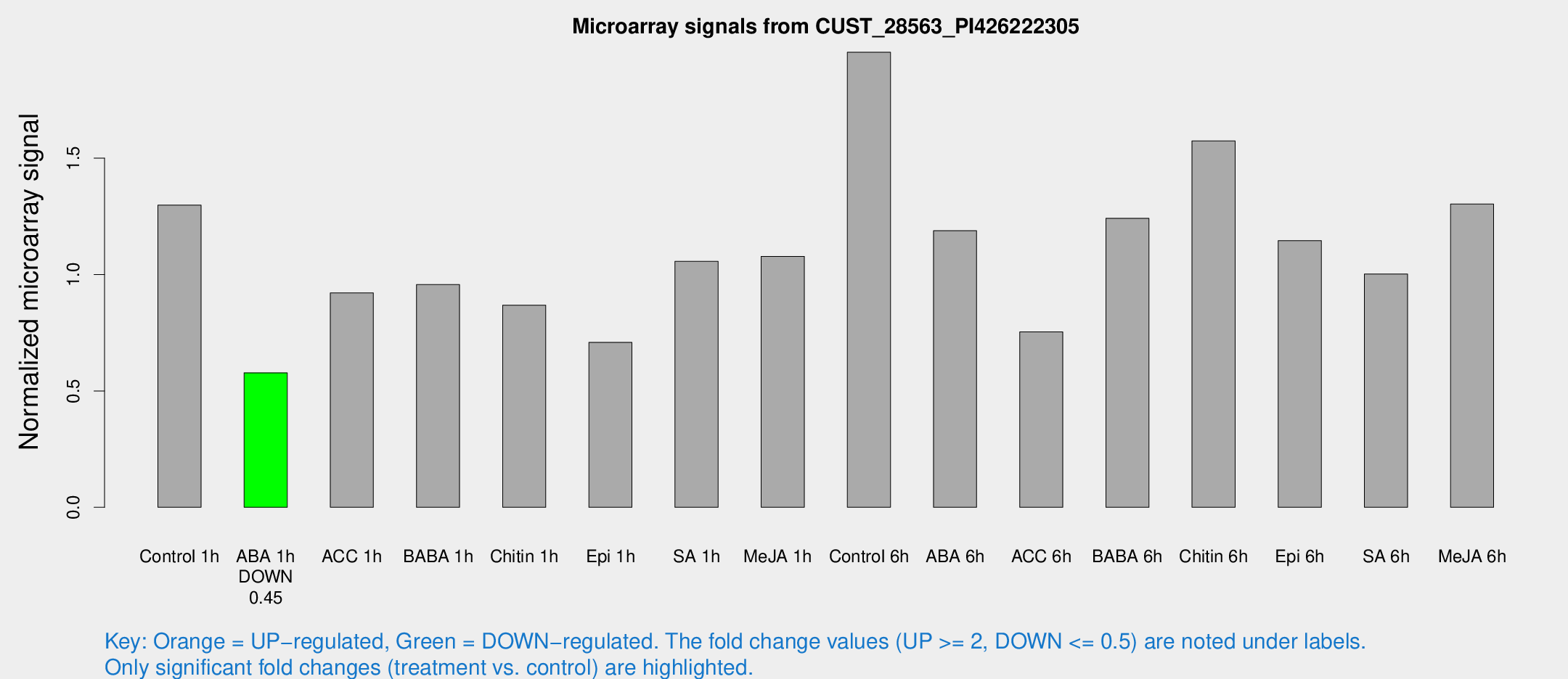
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.4777 | 3.31209 | 1.29843 | 0.322878 |
| ABA 1h | 5.25282 | 3.04775 | 0.577879 | 0.335253 |
| ACC 1h | 9.88648 | 3.6968 | 0.921061 | 0.356891 |
| BABA 1h | 11.6882 | 5.58019 | 0.956698 | 0.437875 |
| Chitin 1h | 8.28269 | 3.23626 | 0.86844 | 0.357172 |
| Epi 1h | 6.36913 | 3.1623 | 0.708617 | 0.362859 |
| SA 1h | 18.6308 | 13.0604 | 1.05661 | 1.28003 |
| Me-JA 1h | 9.73313 | 3.31588 | 1.07805 | 0.422443 |
| Control 6h | 26.9007 | 11.0109 | 1.9549 | 1.24599 |
| ABA 6h | 13.1589 | 3.53099 | 1.18817 | 0.326177 |
| ACC 6h | 9.3744 | 4.03974 | 0.753664 | 0.336934 |
| BABA 6h | 16.6253 | 5.31318 | 1.24132 | 0.541487 |
| Chitin 6h | 20.7369 | 8.04894 | 1.57374 | 0.874285 |
| Epi 6h | 15.3567 | 6.04397 | 1.1458 | 0.505106 |
| SA 6h | 10.312 | 3.53411 | 1.00218 | 0.354008 |
| Me-JA 6h | 16.0424 | 5.99816 | 1.30269 | 0.591709 |
Source Transcript PGSC0003DMT400055510 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36990.1 | +2 | 7e-104 | 340 | 175/293 (60%) | RNApolymerase sigma-subunit F | chr2:15537502-15540016 REVERSE LENGTH=547 |
